International Journal of Marine Science 2015, Vol.5, No.55: 1-9
7
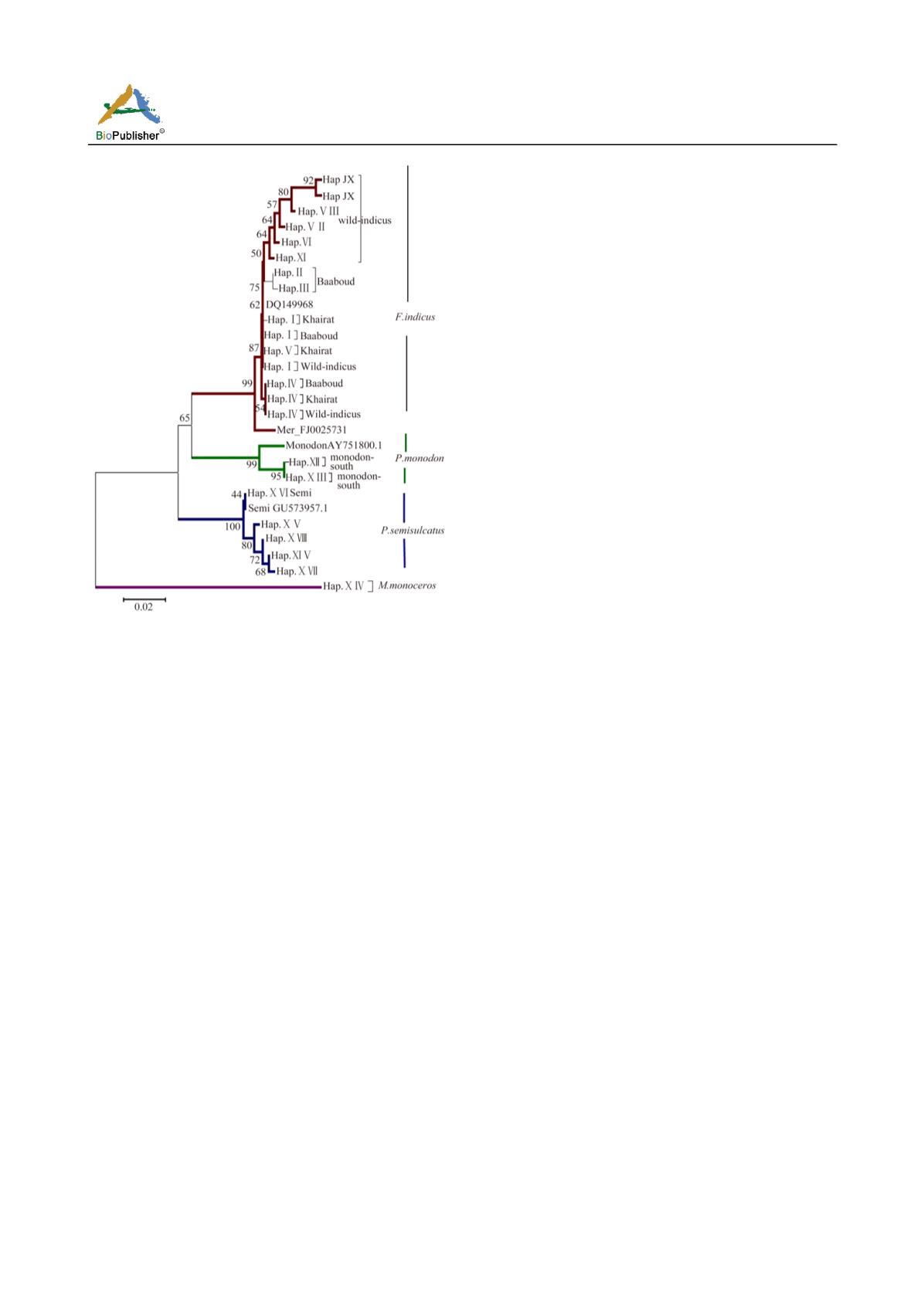
Figure 4 Neighbour-joining tree based on genetic distance
analysis of 16S-rRNA sequences showing the genetic relationships
of
Penaeus
sp. Scale shown refers to genetic distance based on
nucleotide substitutions. Numbers at branching points are bootstrap
support.
M.monoceros
appears as outgroup
highly significant support of boot strap of values. All
the eleven haplotypes of
F. indicus
from the three
location clustered into a single clade. There is a sign
of differentiation between the wild population of
F.
indicus
into one group before it was gathered with
Baaboud and Alkhairat individuals. Their gene bank
reference assigned with them while
F.
merguiensis
(F.J002573.1) form a sister clade with
F. indicus
clade.
Haplotype IV representing at least one individual from
each population grouped in a close position to
F.
merguiensis
(F.J002573.1) considering other species
on the constructed trees, it is obvious that, the 2
haplotypes of
P. monodon
form one clade before
gathering with
P. monodon
(AY751800) retrieved
from gene bank.
The entire five
P
.
semisulcatus
haplotypes cluster
together with their gene bank reference Gu573957.1
clustered into one clade. One of them (Hap.XVI)
seems to be identical with its reference. A forth
distinct clade was formed by the individuals of
M.
monoceros
with clear separation as an out group.
4 Discussion and Conclusions
In conservation genetics, understanding of the
relationship between individuals is particularly important
in captive breeding programmes to reduce incestuous
mating in order to minimize inbreeding and the loss of
genetic variation (Frankham
et al.
, 2002). Knowledge
and studies on genetics can reduce the extinction risk
by helping to develop appropriate population management
programmess that can minimize the risks implied
through inbreeding.
The amplification of 16S rRNA products (480bp)
were in the expected size range as reported by
(Bouchon et al., 1994); (Shekhar et al., 2005) and
Jahmori (2011). While the finfishes investigated by
(Shekhar et al., 2005) have higher size of mitochondrial
segment with respect to the same primers compared to
crustaceans.
For all shrimp species used in the present study, there
was bias towards (A+T) in the base pair composition
16S rRNA gene sequences (68.7). This is commonly
the case in most penaeid species (Baldwin, et al., 1998;
Maggioni et al., 2001). Furthermore, the patterns
described in the present study are consistent with the
description of other arthropod mtDNA sequences
(Crozier et al., 1989; Garcia-Machado et al
.
, 1993) as
well as other marine crustacean mtDNA sequences
(Harrison and Crespi, 1999; Meyran et al., 1997).
Mitochondrial genes such as the large subunit (16S)
ribosomal DNA and Cytochrome C Oxidase subunit I
(COI) are popular markers used in molecular systematic
studies of crustaceans at the species and population
levels (e.g. Baldwin et al.,1998).
Jahmori (2011) stated that; sequences of 16S rRNA
were found to be able to distinguish between 2
morphotypes of Iranian
P. semisulcatus
. whereas (Tsoi
et al., 2005) studied two varieties of
P. japonicus
from
South China Sea. Their results revealed that these
varieties represent distinct clades, with sequence
divergence of about 1% in 16S rRNA gene and 6-7%
in COI gene and 16-19% in the control region, and
they are separated from the other
Penaeus
species.
In the present study haplotype I (Hap.I) and haplotype
IV (Hap.IV) were fairly common between the three
populations of
F. indicus
from Alkhairat, Baaboud and
the wild with Fst values of (0.10957, 0.12459,
0.14817) and gene flow values of Nm (4.06, 3.51, 2.87)


