Cancer Genetics and Epigenetics 2016, Vol.4, No.1, 1-10
3
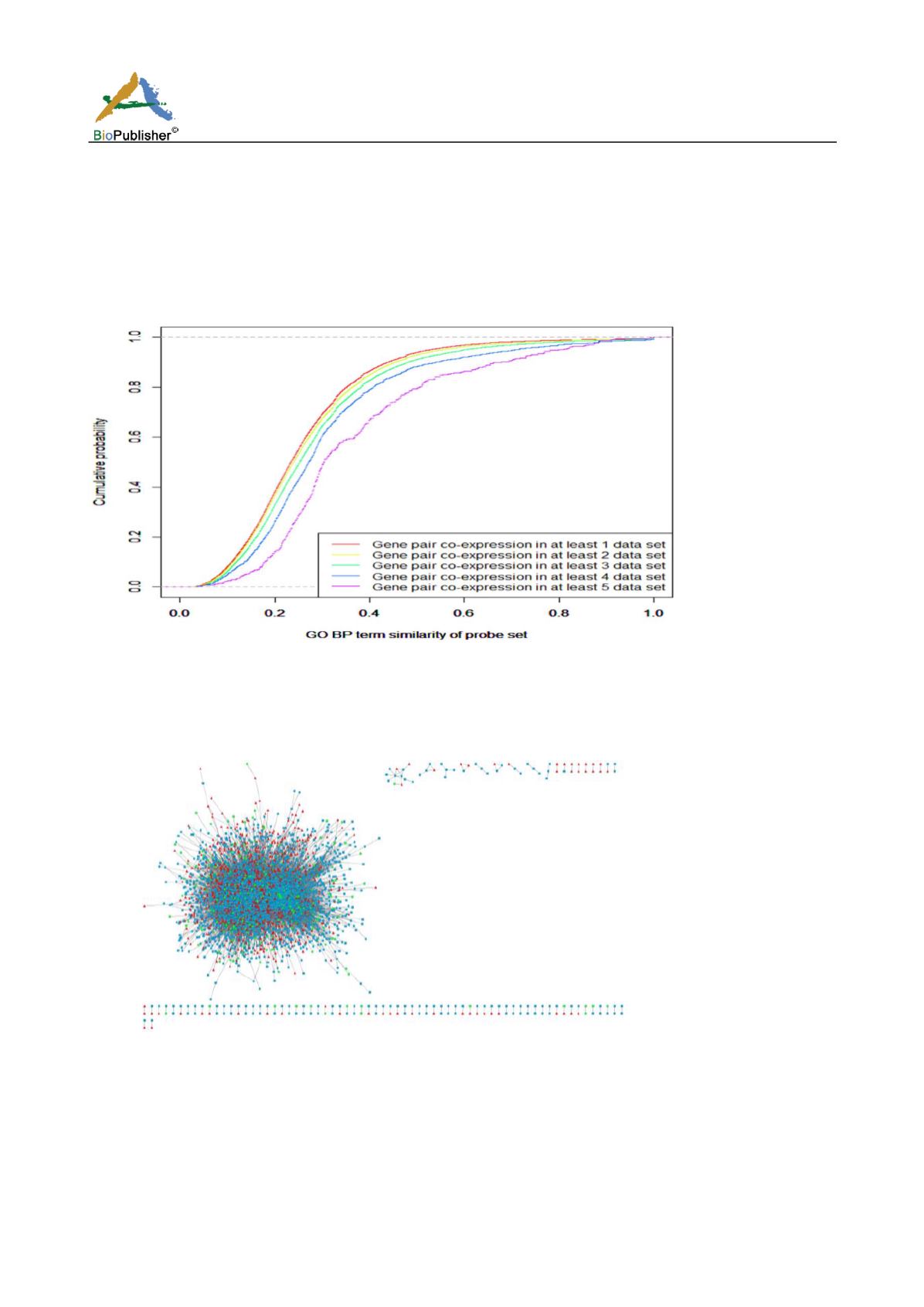
We use GOSemSim package (Yu et al., 2010) of Bioconductor package to calculate the functional similarity of
gene-pairs with same co-expression direction. We found the gene-pair have more similar function, if it has same
co-expression direction in more expression profiles datasets (Figure 2). Since the co-expression genes may be
involved in the same biological processes and have similar functions, in order to ensure that the nodes of
co-expression in the network have more similar functions and the size of the network will not be too small, we
retained gene-pairs which have same direction in more than 3 expression profile datasets to construct
multiple-network.
Figure 2 GO BP term similarity of probe set
The multiple-network (Figure 3) contains 4427 nodes, which include 3370 protein-coding genes (ion channel
protein gene 397, other protein-coding genes 2973) and 1057 ncRNAs. There are 68,647 edges, and 2480
co-expression relationships between ion channel protein genes and ncRNAs.
Figure 3 Multi-Node co-expression networks (Blue nodes represent ion channel protein genes, green nodes represent
protein-coding genes, and red nodes represent non-coding RNAs.)
We analyzed the degree distribution of nodes in the network, found that it obey the power-law distribution of
biomolecular network nodes. Most nodes have small degree value, only small part of nodes has large degree value.
There are 2,885 nodes (65%) that their degree is less than 20, more than half of nodes have less than 10 neighbor
nodes. It indicates that the multiple-network which construct with our method have high reliability.


