Cancer Genetics and Epigenetics 2016, Vol.4, No.1, 1-10
2
mitochondria by control metabolites, which is present in mitochondrial outer membrane and brain postsynaptic
membrane of all eukaryotics. Studies show the expression of VDAC in the frontal cortex and thalamus of
Alzheimer's disease patients (Yoo et al., 2001).
Ion channels and ncRNAs are involved in the occurrence of cancers and neurodegenerative diseases. It is essential
to study relationships between them, but these researches are not sufficient. In this paper, we show a
multiple-network which include ncRNAs, ion channels and others coding-gene. We predicted ncRNAs functions,
and explore the relationship with function of ion channels and ncRNAs. We hope to deepen the understanding of
Alzheimer's disease.
Results
Probe re-annotation of gene expression data sets
After probe re-annotation, there are 133,229 probes mapped to coding-genes and 60,858 probes only mapped to
ncRNAs. To reduce noises and improve the accuracy of probe annotation information, we will map transcripts to
gene, remove probes matching more than one gene and gene contains less than three probes. Finally, we got
22,018 probe-gene pairs, which contain 10,467 genes and 4,216 ncRNAs.
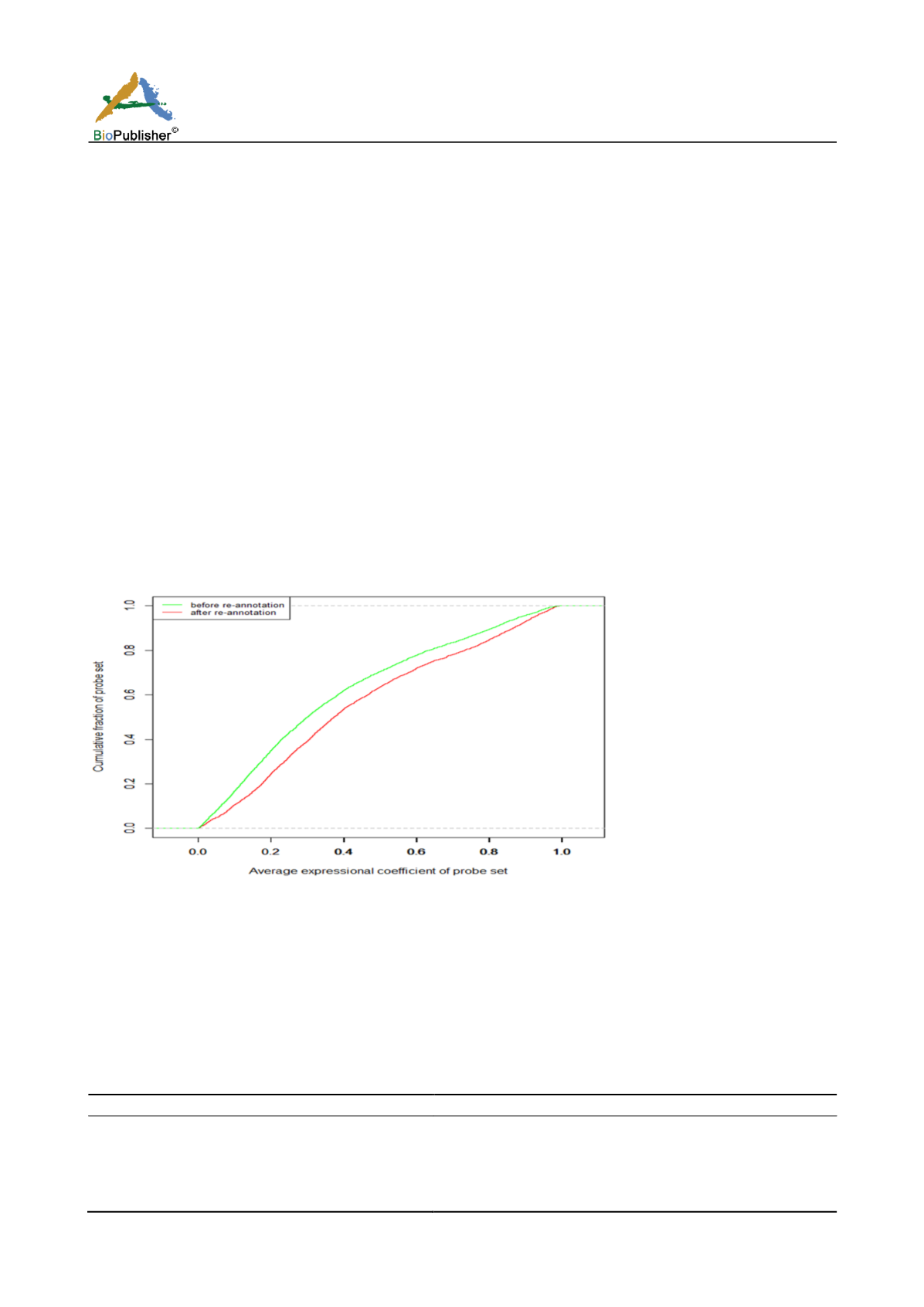
We use GSE48350 to evaluate the accuracy of the result of the probe re-annotation. GSE48350 contains 80
samples; we calculate the mean of correlation coefficient of expression values of probes belongs to the same gene.
We find that after the probe re-annotation, the correlation coefficient of the expression values of the probe of the
same gene is significantly increased (t test, p-value <2.2e-16, Figure 1).
Figure 1 Average expressional coefficient of probe target same coding gene before and after re-annotation.
Multiple co-expression network analysis
We preprocessed each expression profile dataset and calculated co-expression of each gene pair of each
expression profile dataset, respectively. We received a total of 11,413,157 pairs of co-expressed genes. In order to
improve the accuracy of multiple co-expression network, we further analyze the co-expression direction(positive,
negative) of these gene-pairs in the different expression profile datasets, found that only 541 gene-pairs have same
co-expression direction in more than 5 expression profile datasets, and 68,648 gene-pairs have same co-expression
direction in more than 3 expression profile datasets(Table 1).
Table 1 The number of gene pairs with same co-expression direction
The number of expression profile dataset
The number of gene pair of same co-expression direction
1
11413157
2
1264326
3
68647
4
6377
5
541


