Molecular Plant Breeding 2020, Vol.11, No.24, 1-8
4
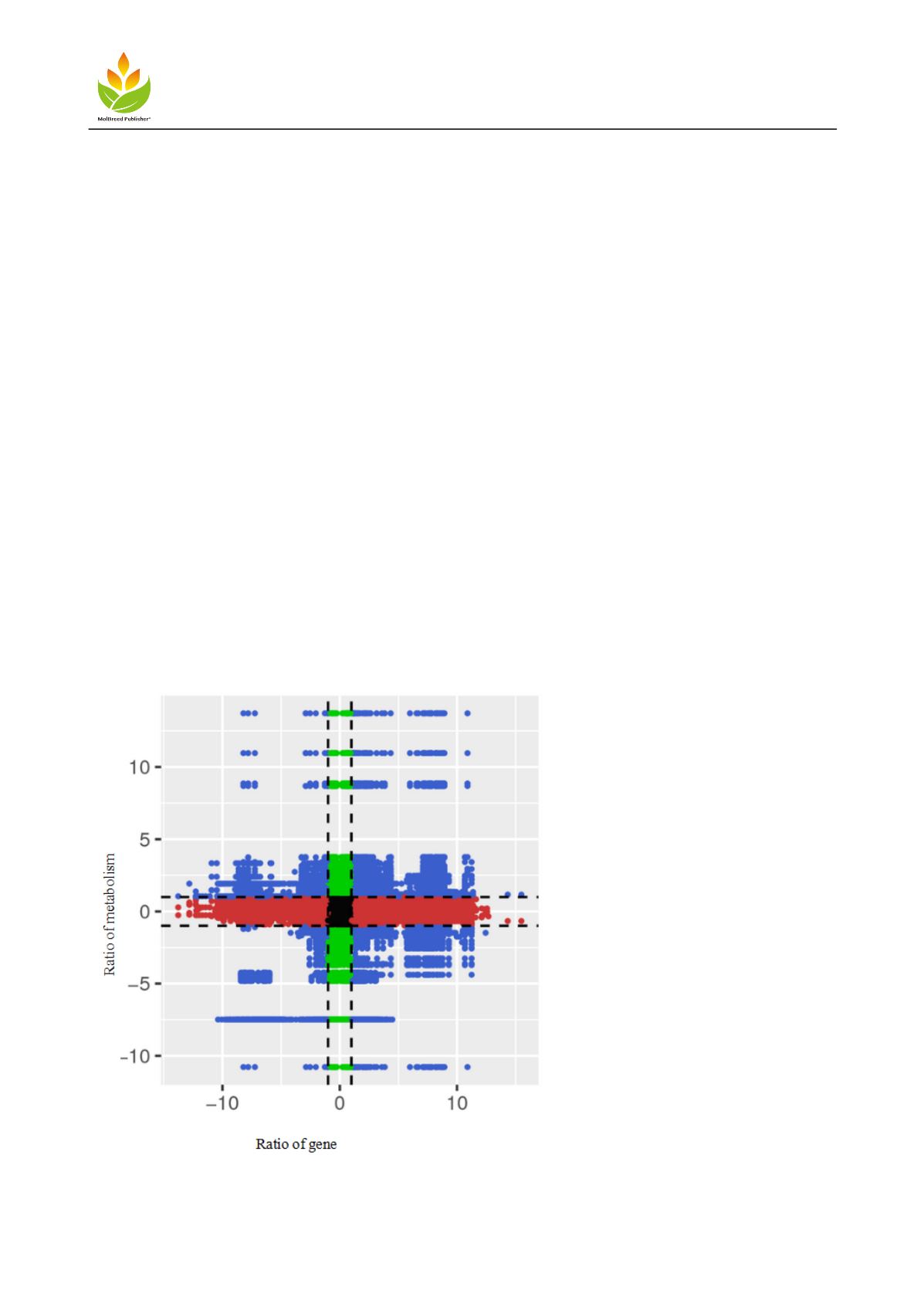
1.4 Correlation analysis between differential genes and different metabolites
The cor program in R was used to calculate the Pearson correlation coefficient of genes and metabolites, and nine
quadrant diagram was used to show the difference multiple of gene metabolites with Pearson correlation
coefficient greater than 0.8 in each difference group. The results are shown in Figure 4: In quadrant 5, there was
no differential expression between genes and metabolites; in quadrants 3 and 7, there was a positive correlation
between gene expression and metabolites, and the change of metabolites might be caused by the positive
regulation of genes; in quadrants 1, 2 and 4, there was correlation between metabolites and gene expression, and
the up regulation of metabolites in quadrants 6, 8 and 9 was negatively correlated with the expression of
metabolites The metabolites were not changed or down regulated (Limin Sun, 2019). According to the correlation
distribution of differential genes and different metabolites, there was significant correlation between differential
genes and differential metabolites, which had a strong regulatory effect on anther metabolism of Yinchuan
Yangjiao male sterile dual-purpose line.
1.5 Analysis of differential genes and metabolites combined with KEGG
Through KEGG enrichment analysis, we can find out the metabolic pathways with significant enrichment of
differential genes and metabolites and show the enrichment degree of pathways with both differential metabolites
and differential genes, so as to further understand the biological functions of differential genes (Kanehisa et al.,
2013). The results showed in Figure 5: the enrichment pvalue value of differential expression genes and
differential metabolites pvalue<0.05, differential expression is significant; however, it is not proportional
distribution in the unified metabolic pathway, which indicates that not all differentially expressed genes regulate
the metabolic pathway. The main metabolites were in Biosynthesis of amino acids, Pyrimidine metabolism,
Tryptophan metabolism and Cysteine, methionine metabolism and Flavonoid biosynthesis. The results showed
that the significantly differentially expressed transcriptional genes played a key role in the above pathways,
resulting in the differential expression of the related products of protein metabolism and flavonoid metabolism in
anthers, which affected the pollen abortion of Yinchuan Zanthoxylum annuum male sterile line.
Figure 4
Nine quadrant of correlation between differential genes and differential metabolites


