International Journal of Horticulture, 2016, Vol.6, No.2, 1-10
4
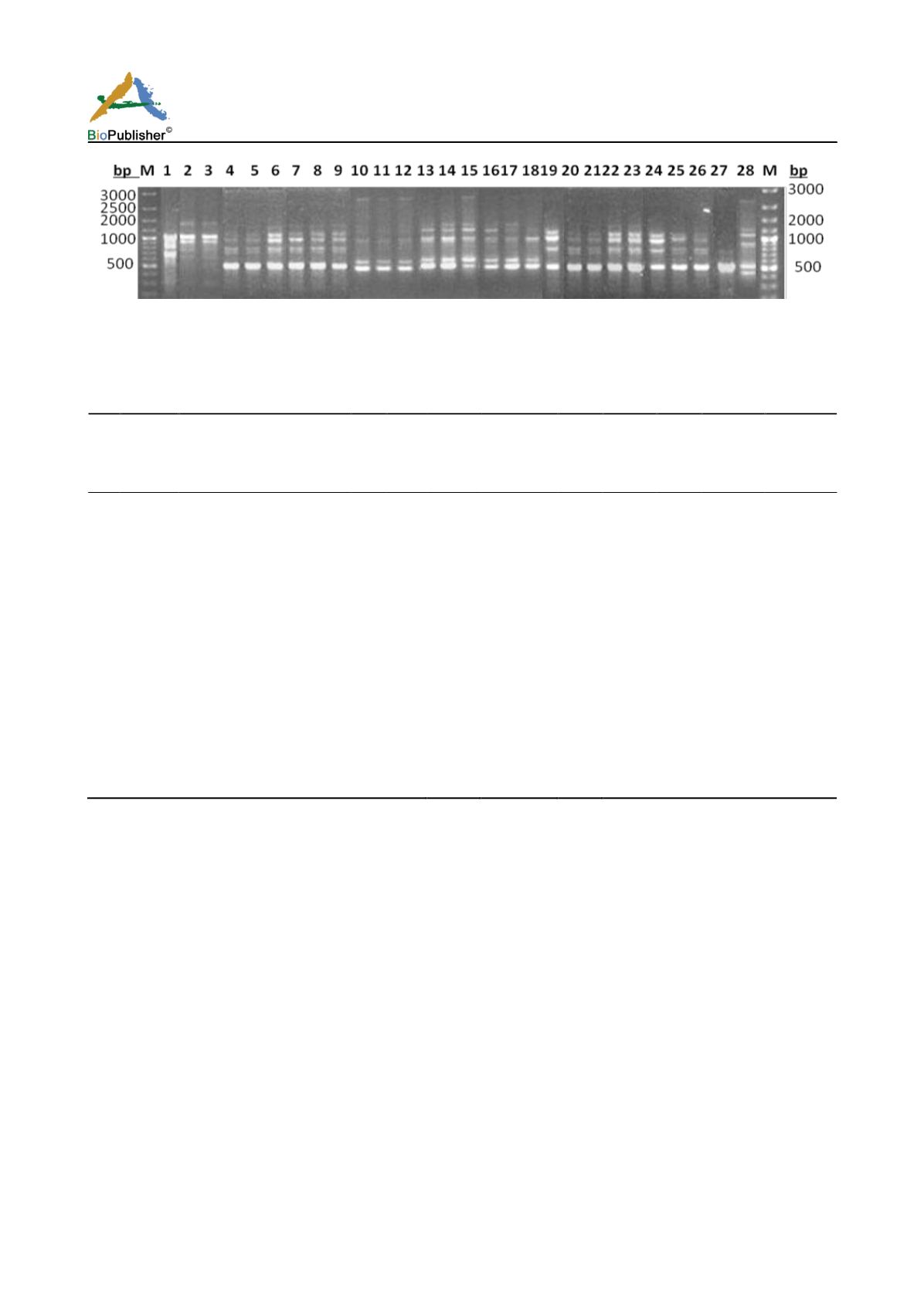
Figure 1c RAPD profiles of eight cashew nut parental genotypes and their 20 promising hybrids amplified with primer OU 34.
M=DNA molecular marker, Lane 1-28 : Parents : RP 1, RP 2, K.pur bold nut, VTH 711/4, Kankady, M 44/3, Vittol 44/3, BPP 30/1;
Hybrids- E 16, B5, A 95, I- 12, F 28, J 13, G 9, A 62, J 6, J 20, C 41, J 12, A 48, C 30, H 8, G 8, A 71, B 27, H 6, D 19
Table 1 Amplified products with different RAPD primers in eight cashew parental genotypes and their 20 promising hybrids
Sl.
No.
Primer
Code
Sequences
(5'-3')
GC
content
(%)
*Tm
(
o
C)
*Tan
(
o
C)
No. of
Poly-m
orphic
bands
No.
of
mono-morp
hic bands
Total
bands
% poly-
mor-phi
sm
PIC
value
Resolvin
g power
(Rp)
Range of
fragment
size ( bp)
1. OU-1
CAGGCGGCGT 80.0
36.0 38.0 6
2
8
75.0
0.58 9.43
480-1830
2. OU-2
ACTGAACGCC 60.0
32.0 34.0 5
0`
5
100.0
0.55 6.57
350-1700
3. OU-3 GGTGAACGCT 60.0
32.0 34.0 6
4
10
60.0
0.31 15.36
680-2800
4. OU-5
AGGACGTGCC 70.0
34.0 36.0 5
0
5
100.0
0.86 3.64
540-2020
5. OU-10 AATCGGGCTG 60.0
32.0 34.0 7
0
7
100.0
0.62 8.21
355-1840
6. OU-16 CTGAGACGGA 60.0
32.0 34.0 7
0
7
100.0
0.73 6.64
520-2100
7. OU-20 AACGTCGAGG 60.0
32.0 34.0 5
1
6
83.3
0.57 7.21
590-2220
8. OU-24 TCCGCTGAGA 60.0
32.0 34.0 8
0
8
100.0
0.68 8.50
660-2500
9. OU-28 TGCTGCAGGT 60.0
32.0 34.0 6
0
6
100.0
0.70 5.50
485-1930
10. OU-29 AACCCGGGAA 60.0
32.0 34.0 9
0
9
100.0
0.58 11.42
390-2850
11. OU 30 CTGCTGGGAC 70.0
34.0 35.0 2
4
6
33.3
0.13 11.14
420-1530
12. OU 32
GAGAGCCAAC 60.0
32.0 34.0 8
1
9
88.8
0.58 10.50
270-1510
13. OU 33
GGCTACACC
60.0
32.0 34.0 2
1
3
66.6
0.47 4.14
750-1280
14. OU 34
AGTCGTCCCC 70.0
34.0 36.0 14
0
14
100.0
0.75 10.21
210-3250
15. OU 35
AGGTTGCAGG 60.0
32.0 34.0 4
0
4
100.0
0.42 5.93
380-1500
Total
94
13
107
As a whole, the total number of bands produced by 15 RAPD primers varied widely among the genotypes. It
ranged from 56 bands in I-12 and J-6 to as high as 72 bands in cashew hybrid E-16. Such differential response of
cashew cultivars was also noted by Samal et al.
(2003a). The data matrix in the present study resolved a total of
1,378 polymorphic amplicons out of 1,742 PCR products across the test genotypes using 15 RAPD primers. This
reveals 79.1% polymorphism indicating higher genetic variation among the hybrids and parents involved. Nine
out of 15 RAPD primers produced 100% polymorphism among which OU-34 revealed highest number of
polymorphic bands (14) of fragment size 210-3250 bp (Figure 1c). Besides, another RAPD primer OU-3 (Figure
1a) amplified ten distinct polymorphic bands (680-2800 bp) followed by Primer OU -29, OU -32, OU-1 and OU
-24 producing 8-9 conspicuous bands each with amplicon size variation 390-2850 bp, 270-1510 bp, 480-1830 bp
and 660-2500 bp respectively. Thus, these primers could be considered highly informative.
GC content, melting temperature (Tm) and annealing temperature are considered most critical factors for
amplification of DNA. In general, melting temperature of primers proportionately increase with the increase in
GC content; and GC-rich primers are expected to yield more polymorphism in the agarose gel. In the present study,
a RAPD primer OU 34 with comparatively high GC content (70%), responded well for production of more
number of amplicons as well as higher level of polymorphism (Table 1). Annealing temperature more than 1-2
°
C
over the melting temperature gave more number of amplicons with good resolution irrespective of the primers
used for amplification.


