International Journal of Horticulture, 2016, Vol.6, No.2, 1-10
3
background without distinct fragments and a few of the primers did not produce any amplified products. As a
result, fifteen RAPD primers were used for genomic DNA amplification of the test genotypes. The list of such
informative primers used in this investigation is presented in Table 1. In the present study, Primer OU-1 amplified
two monomorphic amplicons (1560 bp and 610 bp) while, Primer OU-3 and Primer OU-30 produced four
monomorphic bands each and Primer OU-20, OU-32 and OU-33 each revealed a single monomorphic band of
amplicon size 1950 bp, 1020 bp and 750 bp respectively. A wide array of reproducible PCR products (Figure 1a to
Figure 1c) ranging from 210 to 3250 bp was amplified and it happens to be produced by the primer OU 34 (Figure
1c). Mneney et al. (1998) obtained stable and reproducible RAPD profiles of Tanzania cashew using optimal PCR
reaction conditions. In the present study, the total number of bands ranged from 4-15 with an average of 7.66
bands per primer and the maximum number of bands being produced by primer OU-34. Neto Silva et al. (1995)
observed a total of 27 amplification products with 0-4 bands per primer using six RAPD primers. In the present
investigation, RAPD produced as a whole 94 polymorphic amplicons out of total 107 scorable amplified products
resulting 87.85% polymorphism. Thimmappaiah et al. (2009) generated 60 bands in 100 cashew germplasm
accessions, of which 51 bands were polymorphic (85%) and produced on an average of 5.45 polymorphic bands
per primer. Similarly, Thimmappaiah (2011) assessed 60 accessions of cashew in another study using 20 RAPD
markers that generated 138 bands, of which 8.1 bands were polymorphic per primer. In mango, RAPD primers
yielded 14 monomorphic bands and 96 revealed polymorphism that resulted 87.3% polymorphism as compared to
ISSR 79.38% using 46 cultivars (Bajpai et al.
,
2008), while Samant et al. (2010) obtained 100% polymorphism by
all primers. In this context, 31 bands were reported to be polymorphic out of 120 primers tested in grape (Singh
and Singh, 2011).
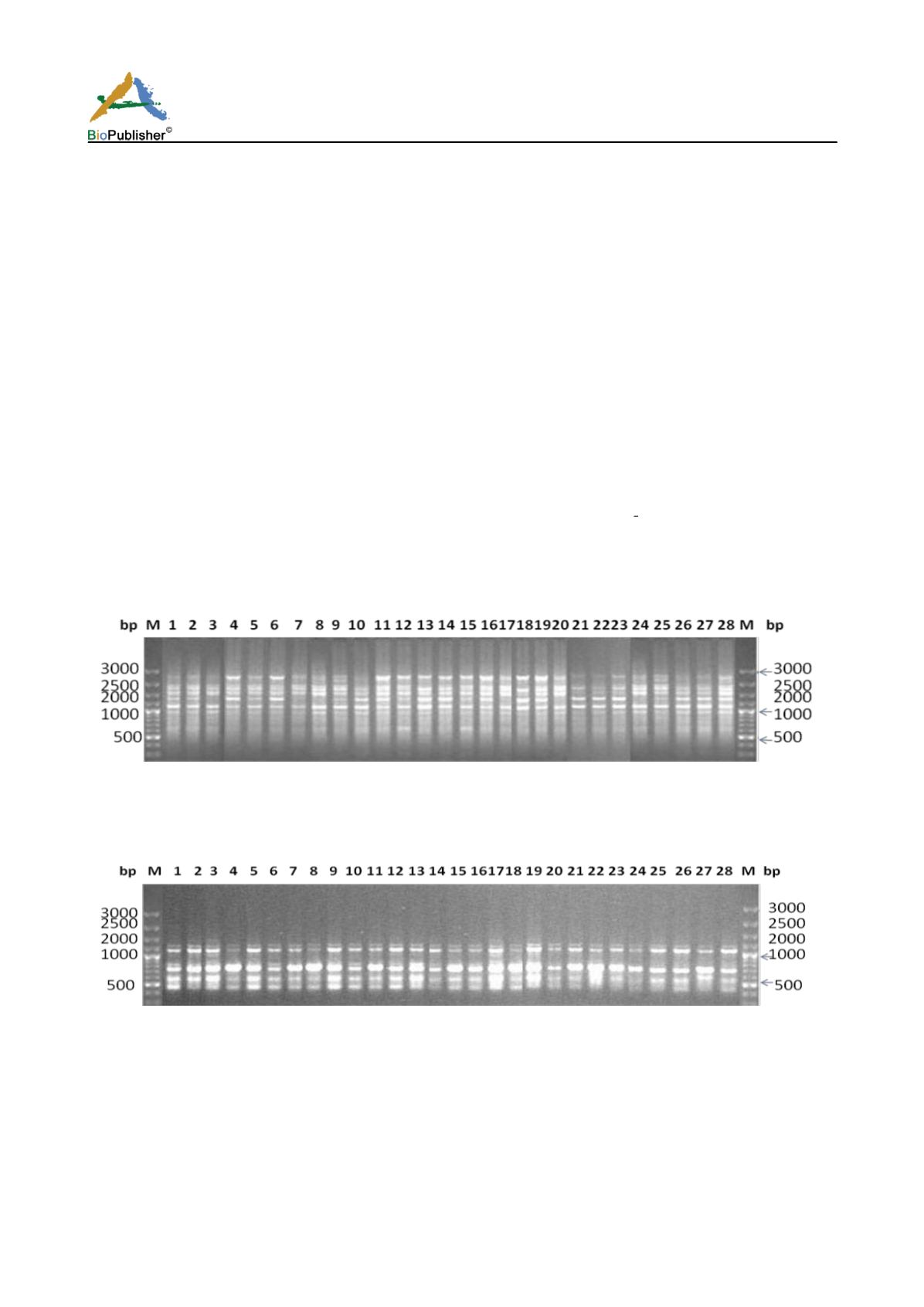
Figure 1a RAPD profiles of eight cashew nut parental genotypes and their 20 promising hybrids amplified with primer OU 3.
M=DNA molecular marker, Lane 1-28 : Parents : RP 1, RP 2, K.pur bold nut, VTH 711/4, Kankady, M 44/3, Vittol 44/3, BPP 30/1;
Hybrids- E 16, B5, A 95, I- 12, F 28, J 13, G 9, A62, J6, J20,C41, J12, A48, C30, H8, G8, A71, B27, H6, D19
Figure 1b RAPD profiles of eight cashew nut parental genotypes and their 20 promising hybrids amplified with primer OU 30.
M=DNA molecular marker, Lane 1-28 : Parents- RP 1, RP 2, K.pur bold nut, VTH 711/4, Kankady, M 44/3, Vittol 44/3, BPP 30/1;
Hybrids- E 16, B5, A 95, I- 12, F 28, J 13, G 9, A 62, J 6, J 20, C 41, J 12, A 48, C 30, H 8, G 8, A 71, B 27, H 6, D 19


