Genomics and Applied Biology 2018, Vol.9, No.10, 62-71
66
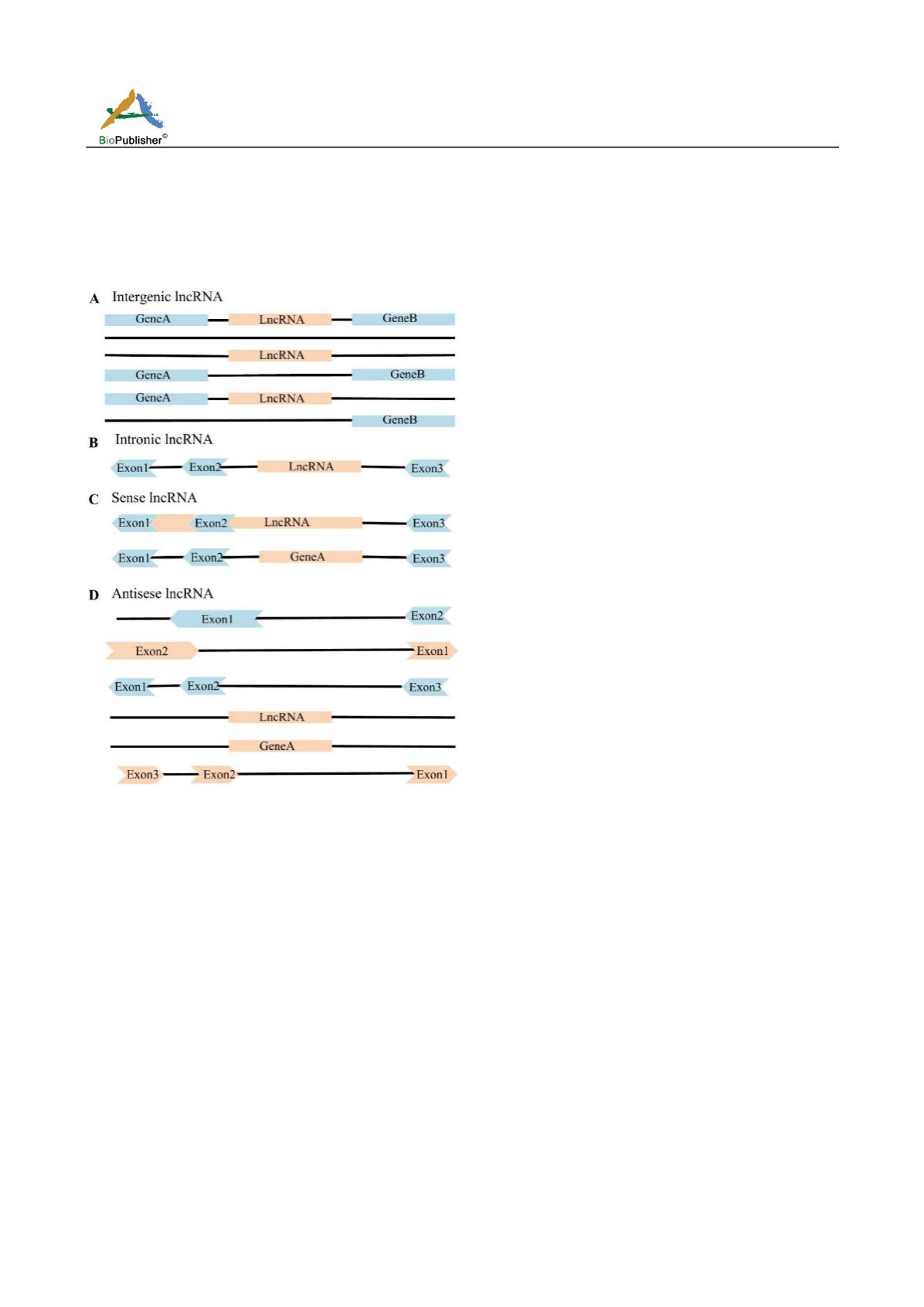
Antisense lncRNA refers to lncRNA that overlaps with one or more exons of antisense chain gene. Intergenic
lncRNA refers to the lncRNA generated by two protein-coding genes. Intronic lncRNA refers to the lncRNA
between two exons from the protein-coding gene. At present, the functions of different types of lncRNA have not
been deeply studied. Finding common features related to the functions of different types of lncRNA will be of
great help to further understand the mechanism of lncRNA in the body.
Figure 3 Classification of lncRNA based on its location on genome
5 Software Related to LncRNA Prediction
The key problem to predict lncRNA is how to distinguish mRNA from ncRNA. At present, the main method to
distinguish mRNA from ncRNA is to establish a classifier, which is mainly based on the sequence characteristics
of lncRNA, such as the modification site of histones, the arrangement of bases and the conservatism of sequences.
One of the representatives of the software developed based on the characteristics of base sequence is PLEK
(predictor of long non-coding RNAs and messenger RNAs based on an improved k-mer scheme) (Li et al., 2014).
This software classifies transcripts based on SVM (support vector machine) algorithm. It calculates the k-mer
frequency of transcripts, and can divide transcripts into protein-coding transcripts and non-protein-coding
transcripts. The classification of PLEK does not depend on sequence alignment or genomic information. In
addition, one of the advantages of the software is that it runs faster. When forecasting the same set of data, the
prediction speed of PLEK software is 8 times that of CNCI and 244 times that of CPC which is the most popular
prediction software. The accuracy of PLEK may fluctuate when it predicts different species. For example, when
using the known mRNA of mice for testing, PLEK wrongly judged the most lncRNA, but if the data of mice were
replaced to the data of corn, PLEK was satisfactory for its higher accuracy performance. In general, it is a stable
and reliable prediction software.
Another classic lncRNA prediction software was CNCI (Coding-non-coding Index), which was developed by the
team of Zhao Yi from the Institute of Computing, Chinese Academy of Sciences (Sun et al., 2013). The software


