![]()
Genomics and Applied Biology 2015, Vol. 6, No. 1, 1-10
http://gab.biopublisher.ca
6
provide valuable information toward a better
understanding of degraded DNA preserved in
postmortem specimens. This information helps to
improve molecular techniques designed to recover and
analyze old DNA to be used for evolutionary studies
and as well as for forensic analysis. Our comparison
of commonly used ancient DNA extraction techniques
based on glass bead-based methods usually cause
noticeable loss of genomic DNA during purification.
We also found that the choice of extraction buffer may
be critical to the success of recovering endogenous
DNA from different types of tissue (for example, soft
tissue, and bone material) preserved under different
physical and chemical conditions. We have obtained
results only either at the lowest or at the highest
amounts of aDNA extracts analyzed. Multiple steps
were taken during DNA amplification procedures to
decrease the effects of PCR inhibitors found in the
amplification reaction. For fossil material, PCR mixes
were set up in dedicated hood in the ancient DNA
laboratory using appropriate contamination control
procedures and then brought to the main molecular
genetics or archaeometry laboratory for thermocyling.
For all ancient and modern reactions, amplification
products were not detected in the negative extraction
(Figure 4 and 5).
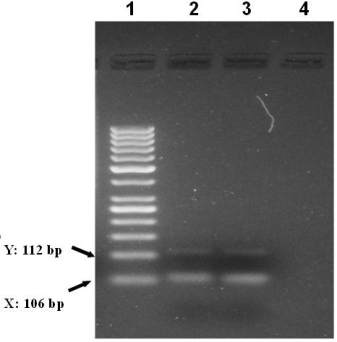
Figure 4 Sex determination based on amelogenin amplification
(Amel A/B) in male and female fossil bones. Amel A/B
indicate PCR fragments spesific to female 106 bp and male
106/112 bp. Agarose gel electrophoresis of PCR products male
and female DNA templates. Lanes 1-4, Lane 1, 50 bp ladder
size standard marker, Lane 2 male and lane 3 female, Lane 4
negative control water blank
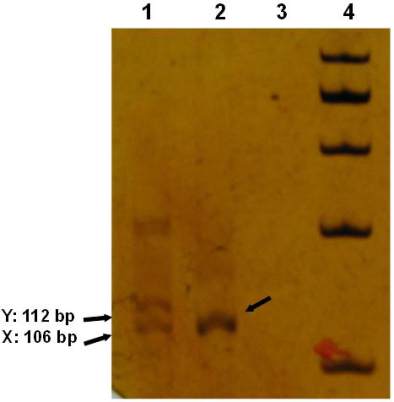
Figure 5 Polyacrilamide gel electrophoresis of PCR products in
male and female fossil DNA templates or 106/112 bp
amelogenin gene PCR products. Respectively, Lanes 1-4, Lane
1: amelogenin male sample 106/112 bp, and lane 2: amelogenin
female PCR fossil sample 106 bp, Lane 3 negative control
water blank or none DNA, Lane 4, 100 bp ladder size standard
marker
1.7 Gel Electrophoresis of PCR Amplicons
PCR product was separated by electrophoresis on 2 %
agarose gel in 1XTAE buffer (45Mm Tris, 1mM
EDTA, pH 8), stained with ethidium bromide. In
addition to electrophoresis of agarose gel, PCR
products were completely loaded in 1.5 %
acrylamide:bisacrylamide gels, stained with Ag(NO
3
)
and agarose gel systems were visualized under UV,
and Poly Acrylamide Gel Electrophoresis systems
were illuminated from above using an white
fluorescent light source (Figure 4, 5). We isolated the
samples from a histological section of the burial place
material and repeated the procedure three times. In
each of the three repeated approaches, amelogenin
could be amplified in all samples showing a
successful DNA extraction (Figure 2). Amplification
products generally showed weak signals in agarose gel
analysis, presumably due to low amount of extracted
material. Nevertheless, high-resolution polyacrylamide
gel electrophoresis demonstrated that the ancient DNA
is derived from a female individual, as in all
amelogenin PCR products only the X-Chromosome
specific 106 bp fragment was visualized (Figure 5).