![]()
Genomics and Applied Biology 2015, Vol. 6, No. 1, 1-10
http://gab.biopublisher.ca
2
analysed to categorise the objects that typically
accompany male and female burials, and anomalies
from the norm can be identified. To do this the
archaeologist needs a reliable method of identifying
the sex of human remains. Current physical
anthropological methods work well with complete
remains, but with incomplete skeletal material,
cremations or juvenile/infant remains the accuracy of
these methods decreases and the element of
subjectivity increases. Much research effort has been
put into the development of a simple, PCR-based test to
identify the sex of human remains. The extraction of
ancient DNA from human remains and the
amplification of sequences from the X and Y
chromosomes in theory should provide a robust and
objective method of identifying the genetic sex of an
individual. In order to identify the gender of ancient
human, typing for a length variation in the X-Y
homologous amelogenin gene (AMEL X and AMEL
Y) (Manucci A., et. al., 1994 and Mitchell R.J., et.al.,
2006). For that reason, DNA sequences spesific to the
X and Y chromosomes may provide an ideal solution.
This sex-typing test is easily performed using the
AMEL 106 and 112 bp primers. In mammals, AMEL
is reported to possess seven exons and two extra exons
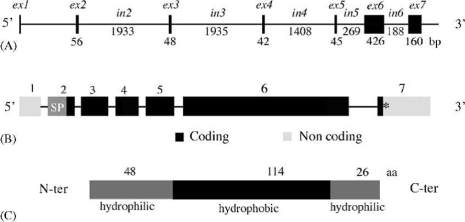
(8 and 9) are found in human (Figure 1). Amelogenin
plays a crucial role in enamel structure and
mineralization, but the function of its various domains
is far to be understood. Evolutionary analysis seems to
be a promising way to approach structure and function
relationships. In this paper, we focus on amelogenin
Figure 1 Amelogenin gene. (A) Gene structure. The five first
exons (ex1-ex5) are small (42-56 bp) and exons 6 (426 bp) and
7 (160 bp) are large. (B) Coding and uncoding exons. Exon 1 is
uncoding, most exon 2 sequence codes the signal peptide, and
the only first three nucleotides of exon 7 are coding for the
protein. (C) Linear representation of the native protein
genes (AMEL) where the amelogenin gene is encoded
on the short arm of both the X (AMELX) and Y
(AMELY) chromosomes in humans. Although
AMELX and AMELY are homologous, they are
different in size and sequence and utilized for the
PCR-based sex test (Manucci A., et. al., 1994).
Genetic difference between male and female is the
presence or absence of chromosome Y. Therefore,
many of existing methods for sex identification are
based exclusively on the detection of sequences
specific for chromosome Y (Bondioli et al. 1989,
Hagelberg E, 1991, Schröder et al. 1990, Wayda et al.
1995). However, the absence of a signal does not
necessarily mean that the sample is of female origin,
as negative results may also be generated by
experimental errors. Thus, the detection of Y- and
X-chromosome specific sequences is advantageous.
Amelogenin gene (AMEL), the origin of which 112
has been traced back to the Precambrian period
(Delgado et al. 2001) has been used as a model in the
field of molecular phylogenetics (Delgado et al. 2005,
Toyosava et al. 1998) as well as in the analyses of
biological traces aiming at determining the sex in
humans and animals (Sullivan et al. 1993, Ennis and
Gallagher 1994, Reklewski et al. 1996, Lechniak and
Cumming 1997, Pfeiffer and Brenig 2005). In forensic
science, designing a single primer pair for the
amelogenin provides the investigation of X and Y
sequences simultaneously for sex identification in
ancient bones. With only a single PCR reaction and
agarose gel electrophoresis, molecular sexing was
reliable and fast. Furthermore, the contamination with
human DNA is sometimes a problem during the
laboratory analysis. However, the amplification of the
human AMEL gene with the same primers resulted in
a different and clearly distinguishable banding pattern.
Our primer design procedure and PCR amplification
provided an excellent tool to DNA from ancient bone.
In conclusion, our study shows how the use of genetic
markers of different mutability might provide an
insight into the history of past necropolises. It also
provides genetic data on ancient Koranza and
Necropal area specimens that could help to confirm or
disprove models developed from modern genetic data
to explain population history. Finally, it provides an
excellent tool to select samples of interest for