![]()
Genomics and Applied Biology 2014, Vol. 5, No. 5, 1-6
http://gab.biopublisher.ca
5
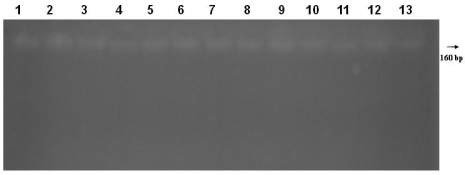
Figure 2 Genomic DNA was isolaled from fossil bone tissue
remains, respectively, Lane 1, 2, 3-13 with Bio Robot EZ1.
aDNA samples submitted to electrophoresis in 1% agarose gel.
Sample codes, respectively, 05BM13, 05BM22, 06BM09,
05BM29, 06BM40, 05BM21, 07BM05, 05BM23, 06BM39,
07BM13, 05BM64, 05BM30, 05BM106 illustrated in the table
1. M: 1 kb ladder size
1.4 Digestion of Fossil DNA by Restriction
Endonucleases
In order to have a better distinguish between product
sizes of AMG gene, the PCR products were digested
by EcoRI/HindIII (Fermantase Life Science)
restriction enzyme.
1.5 Restriction digestion of DNA protocols
Protocol 1 containing; 2 μl 1Xbuffer, 0.5 μl Lambda
EcoRI/HindIII, 0.2 μl 10XBSA, 13 μl water and 2 μl
genomic DNA. 55°C and 2 hours incubation
respectively in lane 2 and 3 identified with protocol 1.
Protocol 2 containing; 2.5 μl 10XBuffer, 0.6μl
Lambda EcoRI/HindIII, 1 μl RNase, 16.9 μl water and
4 μl genomic DNA. 37 °C and 3 hours incubation
respectively in lane 4 and 5 identified with
protocol 2. After digestion, the reaction mixture was
electrophorosed through 0.8% agarose in 50xTAE
buffer. The sample was also tested for nuclease
activity (Figure 3).
1.6 Polymerase Chain Reaction (PCR) Amplification
of Sex Determination
Ancient DNA (aDNA) sex identification was used to
aid in the verification of individual identification
through comparisons to historical documentation of
burials and small sizes human fossil skeletal bones
estimations of sex. The PCR reaction is manipulated
through primer design to favour the amplification of
the Y fragment over the X fragment thus minimizing
the occurrence of ‘false female’ results for male
samples. In this study, the primers for PCR
amplifications used are as follows:
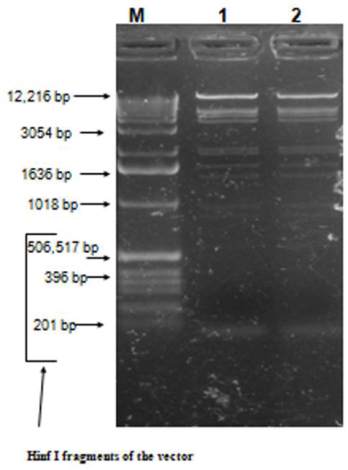
Figure 3 Screenning of agarose gel electrophoresis of fossil
bone DNA digested with restriction endonuclease. M: 1 kb
ladder size standard marker or Hinf I fragments of the vector.
Lane 1-2, genomic DNA isolated from fossil bone by Bio
Robot EZ1 digested with restriction endonuclease
Sequence Amel-A (5’
-
CCCTGGGCTCTGTAAAGA
ATAGTG
-
3’)
Sequence Amel-B (5’
-
ATCAGAGCTTAAACTGGG
AAGCTG
-
3’)
These primers amplify a small region in intron 1 of
the amelogenin gene that encompasses a deletion
polymorphism giving a product of 106 bp for the X
allele and a product of 112 bp for the Y allele, so both
products should be present in males, but only one in
females. 0,5 mg genomic DNA was amplified in a
mixture composed of 5 μL 10XPCR Taq buffer (pH
8.8), 2 mM MgCl
2
, and 10 mM dNTPs (dGTP, dATP,
dTTP, dCTP) at each, 0.5 mM of each primer, and 0.3
U DreamTaq polymerase (Advanced Biotechnologies
Ltd., Fermantase Life Science). Amplification was
submitted to denaturation at 94
o
C for 10 min, 50
amplification cycles with denaturation at 94
o
C for 30
s, annealing at 60
o
C for 10 min and extension at 72
o
C for 1 min in a thermocycler (Biorad, Germany).
PCR blank reactions did not show spot contamination
during the collection of the data (Figure 4, 5). Studies
of ancient DNA from museum and fossil samples can