International Journal of Aquaculture, 2017, Vol.7, No.23, 143-158
152
The GenBank accession numbers of the reference strains are shown besides the names. Horizontal bars in the
dendrogram represent the branch length. Similarity and homology of the neighbouring sequences have been
shown by bootstrap values. Distance matrix was calculated by Tamura-Nei model. The scale bar indicates 0.005
substitutions per nucleotide position.
Bacillus smithii
AB271749.1 served as an out group.
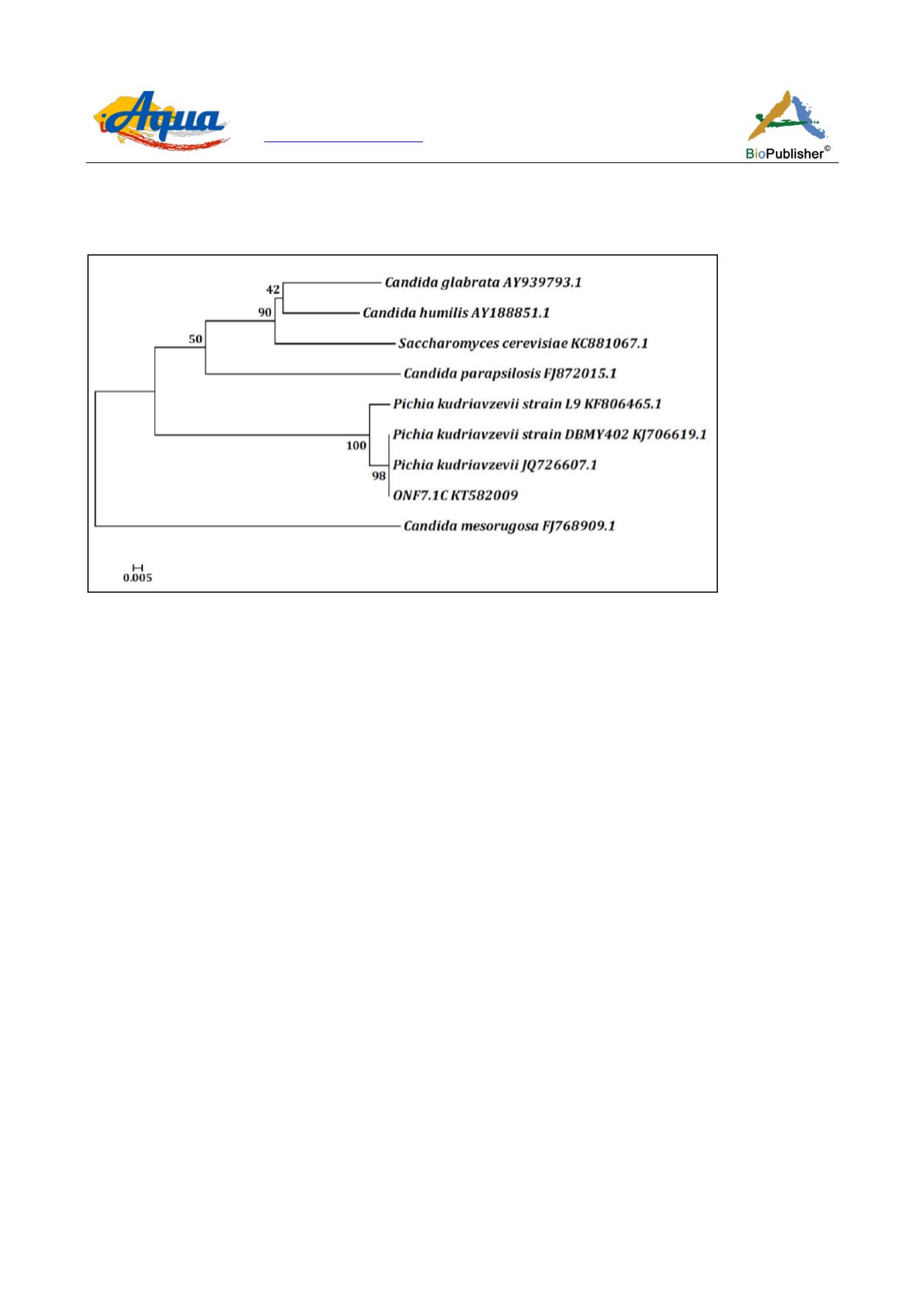
Figure 3 Dendrogram showing phylogenetic relations of the yeast,
Pichia kudriavzevii
ONF7.1C, with other closely related strains
retrieved from NCBI GenBank
The GenBank accession numbers of the reference strains are shown besides the names. Horizontal bars in the
dendrogram represent the branch length. Similarity and homology of the neighbouring sequences have been
shown by bootstrap values. Distance matrix was calculated by Tamura-2 model. The scale bar indicates 0.005
substitutions per nucleotide position.
Candida mesorugosa
FJ768909.1 served as an out group.
4 Discussion
Numerous reports have been published during the last three decades describing bacteria (Ringo et al., 2003; 2007)
and yeasts (Das and Ghosh, 2013; Banerjee and Ghosh, 2014) attached to mucosa and microvilli of the intestine.
Yeasts are ubiquitous microorganisms that grow in various environments where organic substrates are available
(Gatesoupe, 2007). Bacteria are abundant in the environment in which fish live and it is ingested by the fish along
with their diet. Later, they may adapt themselves to the environment of the GI tract and form a symbiotic
association (Saha et al., 2006). Attention has been paid to identify autochthonous fish gut microbiota in order to
gain information on their activities (Cahil, 1990). It could be mentioned that the fish species examined in the
presently reported study were starved for 48 h and their GI tracts were thoroughly washed with sterile chilled 0.9%
saline prior to isolation of microorganisms. Therefore, it may be advocated that the microorganisms isolated in the
present study belong to the autochthonous microbiota as suggested elsewhere (Ray et al., 2010; Ghosh et al., 2010;
Mukherjee et al., 2017). Several investigators have reported the presence of enzyme producing bacteria in the
digestive tract of
Oreochromis
spp. (Sugita et al., 1997; Bairagi et al., 2002; Saha et al., 2006; Ray et al., 2007;
Mondal et al., 2008; Sarkar and Ghosh, 2014; Sasmal and Ray, 2015). The present study is the first one
demonstrating adherent bacteria and yeasts on the gut surfaces of
Oreochromis niloticus
along with antagonistic
properties against the potential fish pathogens. Selection of strong extracellular enzyme producers was considered
as the primary aim of this study. Microbial isolates detected in the present study represented their ability for
production of both, extracellular digestive (amylase, protease, lipase) and degradation enzymes (cellulase,
xylanase, phytase). The observations of Saha and Ray (1998), Ghosh et al. (2002) and Bairagi et al. (2002) were
in agreement with the proposition that fish harbour cellulolytic as well as amylolytic and proteolytic bacteria in
their intestinal tract. Previous study of Saha et al. (2006) revealed that
Bacillus circulans
, isolated from
Oreochromis mossambica
, was able to produce large amount of amylase, protease and cellulase. Two most


