Basic HTML Version


Rice Genomics and Genetics 2012, Vol.3, No.1, 1
-
7
http://rgg.sophiapublisher.com
3
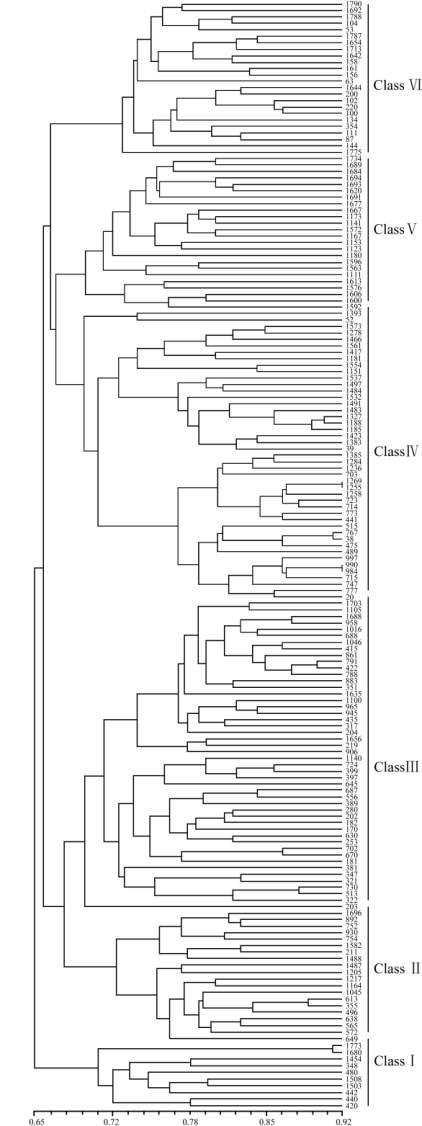
Figure 2 Dendrogram of 171 accessions in the United States
Department of Agriculture (USDA) rice mini-core collection
generated by cluster analysis of genetic distance from 19
OsSPL
primers
Note: Serial numbers are the United States Department of
Agriculture (USDA) rice core collection entries
three-dimensional diagram. The three classification
results were fundamentally consistent; moreover, the
three-dimensional diagram based on the principal
coordinate analysis showed the relationship among the
171 accessions at different directions and levels.
1.5 Global geographical distribution of the subgroups
the 171 accessions
The geographical distribution of the subgroups of the
171 accessions indicated that the accessions of each
subgroup showed strongly similar geographical and
ecological distributions and were not randomly
distributed worldwide. For example, the accessions of
subgroup
Ⅰ
were distributed in tropical rice-growing
areas, with the exception of several accessions that
were distributed throughout China. Accessions of the
same subgroup showed a high frequency in similar
geographical rice-growing areas, such as those in
Panama, Costa Rica, and Honduras, belonging to the
Central American area of subgroup
Ⅵ
.
1.6 Gene-trait association analysis
Based on the results of the cluster analysis, the mean
of the phenotypic data of each subgroup was calculated.
A single sample mean test was analyzed between the
mean of each subgroup and the mean of the phenotypic
data of the total sample, which consisted of the 171
accessions, namely, the gene-trait association.
The gene-trait association (Table 2) showed that there
was a significant difference between the mean of the
spikelets per panicle of subgroup
Ⅰ
and the mean of
the spikelets per panicle of the total sample as well as
a significant difference between the mean of the plant
height of subgroup
Ⅱ
and that of the total sample.
There was also a significant difference between the
mean of the spikelets per panicle of subgroup
Ⅲ
and
that of the sample as well as a significant difference
between the mean of the days to heading of subgroup
Ⅵ
and that of the sample. The mean of the plant height
of subgroup
Ⅴ
and that of the sample almost reached
a significant level, with a difference probability of
0.052 (P=0.052). There was no significant difference
between the mean of each trait of subgroup
Ⅳ
and
that of the sample, which indicated that the mean of
each trait of subgroup
Ⅳ
was no offset. There was no
significant difference in the mean of the tillers per

