Basic HTML Version


Rice Genomics and Genetics 2012, Vol.3, No.1, 1
-
7
http://rgg.sophiapublisher.com
2
expression has been confirmed in rice (Xie et al., 2006).
In this study, 19 pairs of
OsSPL
gene primers were
designed according to their sequences and used for a
polymorphism analysis of 171 accessions (Agrama et
al., 2009) in a United States Department of Agriculture
(USDA) rice mini-core collection established in the
US. We analyzed the gene-trait association between
the genotypes and phenotypes to elucidate the association
between
OsSPL
gene polymorphisms and the major
agronomic traits.
1 Results and Analysis
1.1 Global distribution of the 171 accessions and
their composition
The 171 accessions in the USDA rice mini-core
collection consist of 163 Asian cultivars (
Oryza sativa
),
7 African cultivars (
Oryza glaberrima
), and 1 common
wild rice (
Oryza rufipogon
griff.); these cultivars are
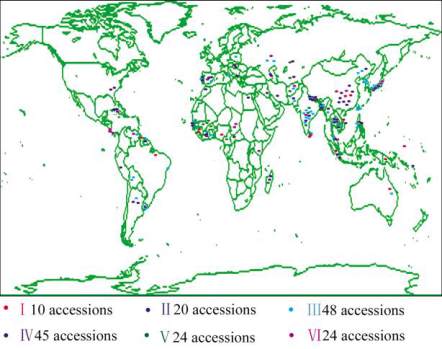
distributed throughout 68 countries (Figure 1). There
are 93 accessions in Asia (~54.4%), with 19 China's
accessions, the most among all of the countries, and
13 India's accessions (the second most). Other countries
in globally have 1~3 accessions.
Figure 1 Global distribution of 171 accessions and six subgroups
of the USDA rice mini-core collection
Note:
Ⅰ
,
Ⅱ
,
Ⅲ
,
Ⅳ
,
Ⅴ
and
Ⅵ
represents one subgroup,
respectively
1.2 Sequence analysis between the Japonica and
Indicia
OsSPL
genes
Single nucleotide mutations and/or cantlet-base dele-
tions were identified between the
Keng
(
Japonica
)
Nipponbare
OsSPL
genes and
Hsien
(
Indica
) 9311
OsSPL
DNA sequences based on http://www.gramene.
org/Multi/blastview/BLA_K8FkdkRLV. A pairwise
protein alignment showed that the identity in protein
sequences of the
Keng
(
Japonica
) Nipponbare
OsSPL
genes and
Hsien
(
Indica
) 9311
OsSPL
genes was
100%, which indicated no difference between the
coding sequences of the
Keng
(
Japonica
) Nipponbare
and
Hsien
(
Indica
) 9311
OsSPL
genes.
1.3 PCR assay of 171 accessions
In this study, 19 pairs of
OsSPL
gene primers were
designed, and 140 bands were amplified totally. Among
them, 128 were polymorphic, comprising a majority
(up to 91.4%) of the mini-core collection. The average
number of alleles per locus was 7.4 (Table 1).
1.4 Cluster and principle coordinate analysis
Using the unweighted pair group method with
arithmetic mean (UPGMA), our cluster analysis based
on the PCR results agreeably classified the 171
accessions into two groups when using a simple
matching coefficient of 0.65. The first group consisted
of 10 accessions, and the second group consisted of
161 accessions. At the simple matching coefficient of
0.687, the first group comprised one subgroup
Ⅰ
,
and the second group was divided into five subgroups
(Figure 2). Subgroups
Ⅱ-Ⅵ
consisted of 20, 48, 45,
24, and 24 accessions, respectively. The 7 African
cultivars were classified into three subgroups: USDA
IDs 321 and 1688 belonged to subgroup
Ⅲ
, USDA
IDs 1689, 1691, 1693, and 1694 were categorized into
s
ubgroup
Ⅴ
, and USDA ID 1692 was put in
subgroup
Ⅵ
. Common wild rice (USDA ID 1703)
was classified into the
Ⅲ
subgroup. Thus, there was
no obvious differentiation between the Asian and
African cultivars in the
OsSPL
gene family.
The results of the principal coordinates analysis based
on the original matrix of the PCR amplification
indicated that the first two and first three coordinates
could explain 70.21% and 72.17% of their correlation,
respectively. Two-dimensional and three-dimensional
scatter plots of the 171 accessions were generated
(Figure 3; Figure 4), showing a strip-type discrete
distribution in the two-dimensional diagram that was
clearly divided into two groups and six subgroups;
these groupings were more clearly demonstrated in the

