Molecular Plant Breeding 2015, Vol.6, No.19, 1
-
7
3
Table 3 Pair-wise Fst value among
Dracocephalum thymiflorum
populations. (Upper diagonal = Fst value, lower diagonal = P value).
Pop1
Pop2
Pop3
Pop4
Pop5
Pop1
--
0.151
0.216
0.128
0.157
Pop2
0.001
--
0.429
0.306
0.429
Pop3
0.001
0.001
--
0.228
0.025
Pop4
0.001
0.001
0.001
--
0.194
Pop5
0.007
0.001
0.242
0.001
--
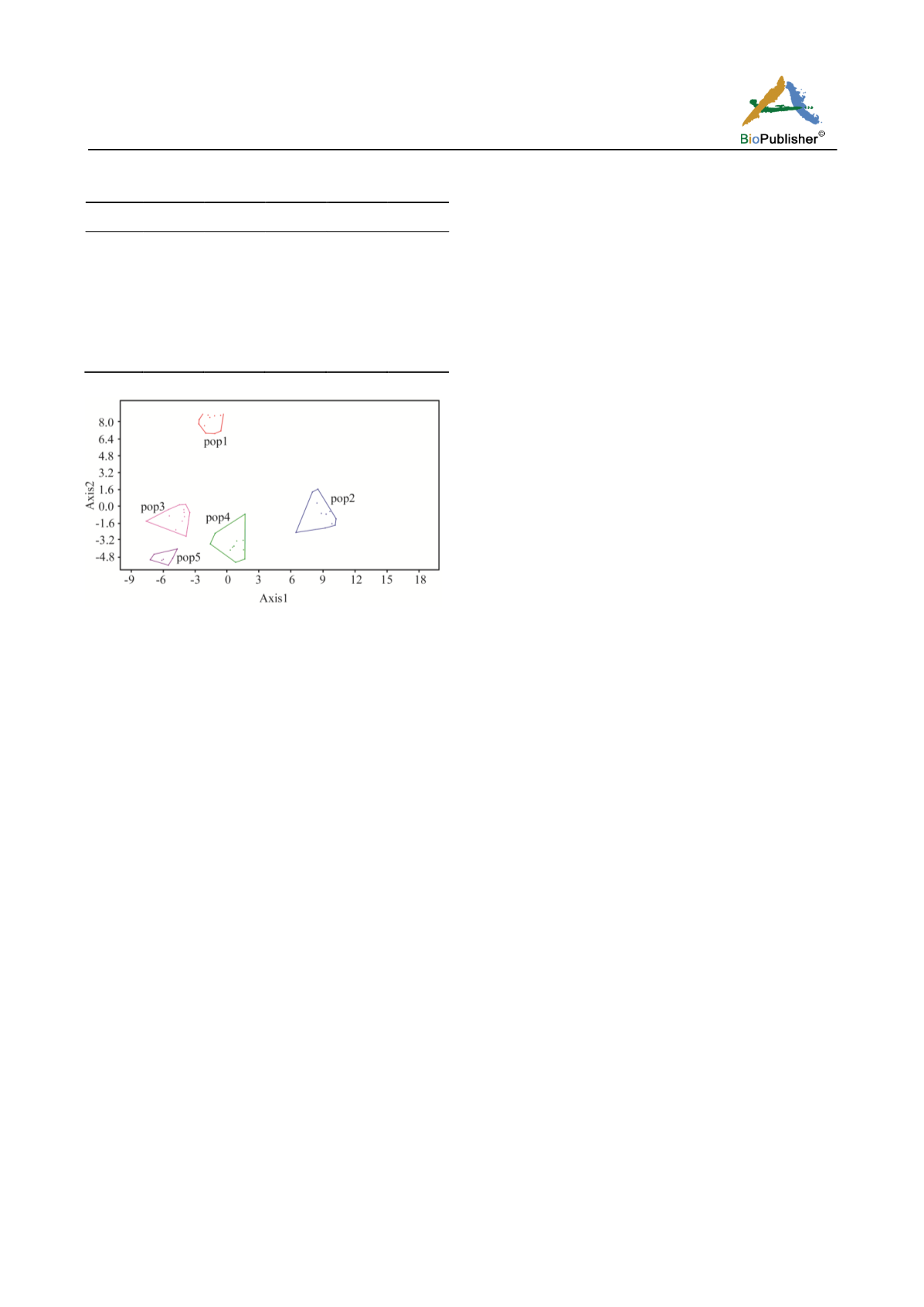
Figure 1 CVA plot of
Dracocephalum thymiflorum
populations
1.3 Population genetic structure and gene flow
The Evanno test and K-Means clustering produced the
best number of genetic groups as K = 2. Therefore,
STRUCTURE analysis was performed for K = 2. The
STRUCTURE plot obtained (Figure 4) revealed close
genetic affinity between the studied populations due to
ancestral shared alleles. However, it also revealed that
population 2 is genetically differentiated in its genetic
structure. The plot also showed a higher degree of
genetic admixture among populations 3-5, followed
by population 1. This Bayesian approach analysis is in
agreement with PCoA plot result.
Population assignment test revealed more detailed
information on genetic admixture and gene flow
among these populations (Table 4). This test is based
on maximum likelihood of plants membership to their
own or other populations. It revealed that out of 55
studied plants, 16 plants were inferred to be from other
populations. The population 2 was comparatively less
involved in gene exchange with the other populations
and the main stream of gene flow occurred between
populations 1, 3, 4, and 5.
The Mantel test produced significant correlation (r =
0.41, P = 0.01) between genetic distance and
geographical distance of the studied populations.
Therefore, the populations that are in closer vicinity
had the chance for gene flow between each other.
LFMM analysis identified 12 out of 46 ISSR loci are
adaptive.
2 Discussion
With increase in the size of human population, the
crop plants and medicinally important plant taxa are
consumed and destroyed faster that before. This treat
is greater for those plant species that are rare and grow
in limited number and therefore, their conservation
become an important task (Sheidai et al., 2013, 2014).
Medicinal plants such as
Dracocephalum thymiflorum
are extensively used by locals and therefore are
subject to be reduced in number or elimination from
the natural habitat. The disappearance and fragmentation
of natural populations could lead to reductions in the
rate of gene flow among populations. This in turn
increases in genetic differentiation among populations
and reductions in genetic variation within populations
due to genetic drift (Setsuko et al., 2007; Hou and Lou,
2011).
To plan a properly oriented conservation plan, the
knowledge of genetic diversity available in the target
species becomes important. The genetic variability
can help plant taxa to adapt to changing environments
they are growing in (Freeland et al., 2011).
The AMOVA test indicated that out of total genetic
variation, 75% was due to within population genetic
variability in the studied
Dracocephalum thymiflorum
populations. This should be related to outcrossing
nature of this plant species. The presence of high
within population genetic variability helps the population
to cope with local environmental changes.
STRUCTURE analysis and K-Means clustering revealed
some degree of population genetic fragmentation in the
studied populations. This was mainly due to the plants
in population 2 that differed genetically from the others.
Moreover, AMOVA, Gst and differentiation parameters
revealed significant genetic difference among the studied
populations. Among population genetic differentiation is
the product of absence and or limited between
population gene flow, genetic drift, inbreeding, and local
adaptation (Hou and Lou, 2011; Sheidai et al., 2014).
The assessments of the levels of within- and among-


