Molecular Plant Breeding 2015, Vol.6, No.19, 1
-
7
5
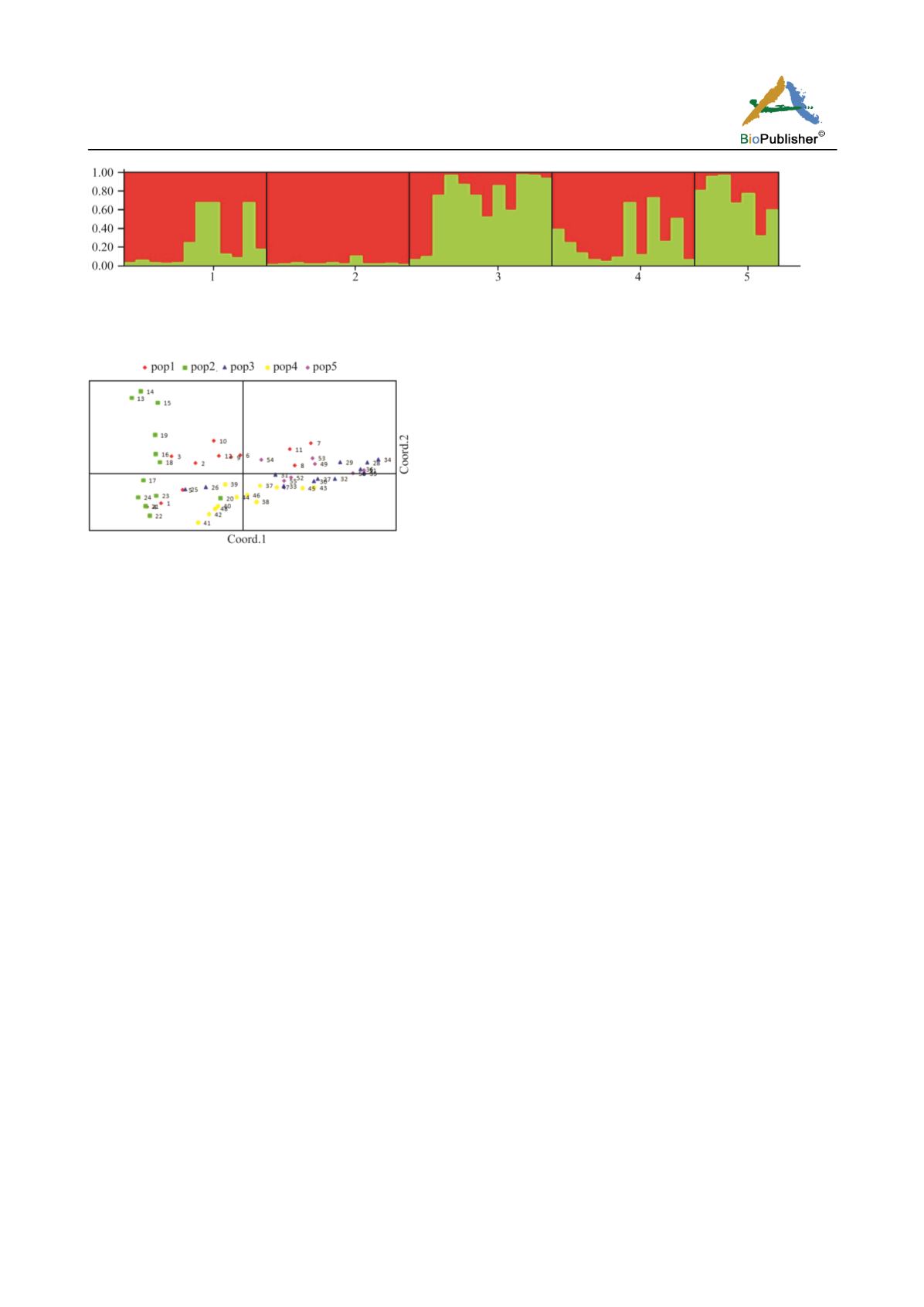
Figure 4 STRUCTURE plot of
Dracocephalum thymiflorum
populations. (Populations 1-5 are according to Table 1)
Figure 3 PCoAplot of
Dracocephalum thymiflorum
populations.
(Populations 1-5 are according to Table 1).
Gene flow is also important in conservation contexts,
particularly for the species with few local populations.
In these species, the genetic characteristics are strongly
influenced by genetic drift and inbreeding (Frankham
et al., 2002). Fortunately, although
Dracocephalum
thymiflorum
populations are few in number and are
confined to some ecological places, they showed good
within-population genetic variability and limited amount
of among population gene flow. Gene flow among
local populations could mitigate losses of genetic
variation caused by genetic drift in local populations
and thus save them from extinction (Richards, 2000).
Mantel test revealed isolation by distance in the
studied
Dracocephalum thymiflorum
populations. The
plant species that form geographical populations, as
geographical isolation increases, a reduction in both
seed dispersal and pollen flow will result in decreased
gene flow between populations. The resulting genetic
isolation may lead to pronounced geographical structuring
in genetic variation within a species as population
differentiation increases (Jump et al., 2003). High
degree of genetic variability observed in
Dracocephalum thymiflorum
populations can be used
in programming conservation plan of this medicinal
plant species in Iran.
We conclude that, genetic differentiation, genetic drift,
limited gene flow and local adaptation have played
role in
Dracocephalum thymiflorum
population
divergence.
3 Materials and methods
3.1 Plant materials
Fifty-five plant specimens were randomly collected
from 5 geographical populations of
Dracocephalum
thymiflorum
L. Details of localities are provided in
Table 1. Voucher specimens are deposited in Herbarium
of Shahid Beheshti University (HSBU). Fresh leaves
were collected and used for DNA extraction and
molecular study.
3.2 DNA extraction and ISSR assay
Fresh leaves were collected randomly in each of the
studied populations and dried in silica gel powder.
Genomic DNA was extracted using CTAB activated
charcoal protocol (Sheidai et al., 2013). The quality of
extracted DNA was examined by running on 0.8%
agarose gel.
Ten ISSR primers; (AGC)5GT, (CA)7GT, (AGC)5GG,
UBC810, (CA)7AT, (GA)9C, UBC807, UBC811,
(GA)9T and (GT)7CA commercialized by UBC (the
University of British Columbia) were used. PCR
reactions were performed in a 25 μl volume containing
10 mM Tris-HCl buffer at pH 8; 50 mM KCl; 1.5 mM
MgCl
2
; 0.2 mM of each dNTP (Bioron, Germany); 0.2
μM of a single primer; 20 ng genomic DNA and 3 U
of
Taq
DNA polymerase (Bioron, Germany). The
amplifications
,
reactions were performed in Techne
thermocycler (Germany) with the following program:
10 Min initial denaturation step 94°C, 30 S at 94°C; 1
Min at 57°C and 1 Min at 72°C. The reaction was
completed by final extension step of 7 Min at 72°C.
The amplification products were visualized by running
on 2% agarose gel, followed by the ethidium bromide
staining. The fragment size was estimated by using a
100 bp molecular size ladder (Fermentas, Germany).


