Basic HTML Version

Molecular Pathogens
6
For the isolated
Vibrios sp
, the highest (2.00 nm) and
lowest growth (0.17 nm) range was observed from
V.mimicus
and
V.paraheamolyticus
based on various
temperature (20
℃
~
60
℃
).
V.cholera
and
V.vulnificus
exhibited the lowest (0.00 nm) and highest (2.00)
range in different salinity (5%~25%). Effect the
different carbon source (Glucose, Sucrose and Lactose)
on isolated vibrios sp. the lowest (0.02 nm) and
highest (1.32 nm) value were observed in
V.mimicus
and
v.alginolyticus.
For the isolated
Vibrio sp.
at various
pH levels (6~10),
V.paraheamolyticus
and
V.mimicus
are
expressed to lowest (0.02) and hight range (1.09).
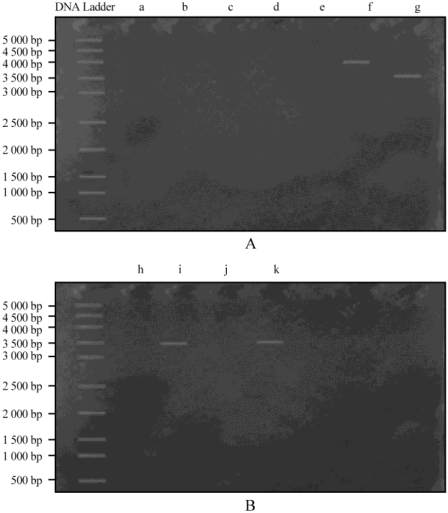
The plasmid profiling revealed that out of 11
isolates analysed for plasmid only 4 isolates
V.parahaemolyticus
(2),
V.vulnificus
(1)
, V. alginolyticus
)
and
their molecular weight compared with 5000bp
DNA ladder and their molecular weight identified in
4000bp and 3500bp, while the other 7 isolates were
without plasmids that are represented in Figure 1, 2
and Table 6.
Figure 1 Fishes in this study
Note: A: Local name: ooli; English name: shela; Biological
name: Sphyraena barracuda; B: Local name: nagarai; English
name: indian goat fish; Biological name: Upeneus sp.; C: Local
name: ayalai; English name: mackral; Biological name:
Rastrelliger kanagurta; D: Local name: chalai; Biological name:
Sardinella sp.
3 Discussion
Tanil et al
(2005) proceeded to study Twenty-one
Vibrio parahaemolyticus
isolates representing 21
samples from coastal seawater from three beaches in
peninsular Malaysia which were found to be sensitive
to streptomycin, norfloxacin and chloramphenicol.
Resistance was observed to penicillin (100%),
ampicillin (95.2%), carbenicilin (95.2%), erythromycin
(95.2%), bacitracin (71.4%), cephalothin (28.6%),
moxalactam (28.6%), kanamycin (19.1%), tetracycline
(14.3%), nalidixic acid (9.5%) and gentamicin (9.5%).
Plasmids of 2.6 to 35.8 mDa were detected among
plasmid-containing isolates.
Ramalingam and Shyamala (2006) reported
V. cholerae
isolates were subjected to 11 antibiotics using disc
Figure 2: Gel image of plasmid profiles of the randomly
selected
Note: A: a-V.parahaemolyticus, b-V.parahaemolyticus, c-V.vulnificus,
d-V.cholerae, e-V.alginolyticus, f-V.paraheamolyticus; B:
h-V.cholerae., i-V.parahaemolyticus, j-V.mimicus, k-V.vulnificus
Table 6 Plasmid formation of randamly selected the isolated
Vibrios
from marketed sea food samples
DNA
a
b
c
d
E
f
g
h
i
J
k
RESULT
-
-
-
-
-
+
+
-
+
-
+
Note: (+) – Positive, (
-
) – Negative, a
-
V.parahaemolyticus
, b
-
V.parahaemolyticus
, c
-
V.vulnificus
, d
-
V.cholerae
, e
-
V.alginolyticus
,
f
-
V.paraheamolyicust
, g
-
V. parahaemolyticus
,h
-
V cholerae.
, i
-
V.parahaemolyticus
,j
-
V.mimicus
, k
-
V. vulnificus

