Basic HTML Version


Journal of Mosquito Research, 2013, Vol.3, No.4, 21
-
32
ISSN 1927-646X
http://jmr.sophiapublisher.com
26
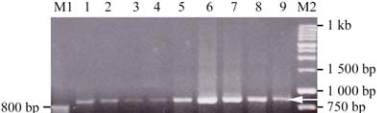
Figure 4 Ethidium bromide stained agarose gel of PCR
amplified fragments using degenerate primers for insect
transferrin gene
Note: M1: 50 bp DNA Ladder marker; M2: Molecular size
marker: G5711, 1 kb DNA Ladder, promega#274214; Lanes 1,
2, 3, 4:
H. bacteriophora
treatment after 3, 6, 9, 12 hr,
respectively; Lane 5: control, Lanes 6, 7, 8, 9:
S. carpocapsae
treatment after 12, 9, 6, 3 hr, respectively. The arrow about 858 bp
shows the
Cx. quiuquefasciatus
PCR amplified fragmet
The band intensity was detected by determination of its
integrated display value (IDV) using gel documentation
system (Alfainnotech gel documentation), Table 1. By
increasing the incubation time from 3 to 12 hr, the
IDV increased when using SC nematodes. In the same
time it is worthy to mention that the least intense band,
corresponding to insects exposed to nematodes for a
period of only 3 hours, was lower than those of the
control group. On the other hand, those mosquito
larvae that were treated with
H. bacteriophora
nematodes showed the down-regulation of the
transferrin gene as shown from the decreasing IDV
than control group by increasing incubation period
from 3 to 12 hr.
3.3 Cloning and sequencing of Tsf coding region
The amplified Tsf coding sequence was ligated into
pGEM-T cloning vector (Invitrogen) by TA ligation
procedure, following the manufacturer’s instructions.
The ligation product was used to transform The
Mach1™- T1
®
E. coli
chemically competent cells.
Successfully transformed cells were selected by blue/
white selection, where white colonies were selected.
Plasmid mini preparation was prepared and double
digested with
Bam
HI/
Hin
dIII. Sequencing and
blasting of the cloned Tsf (Figure 5) was inserted
against non-redundant nucleotide database of NCBI
(http://blast.ncbi.nlm.nih.gov/blast/Blast.cgi?CMD=W
eb&PAGE_TYPE=BlastHome), which showed that
the amplified sequence is the coding sequence of Tsf
gene.
Figure 5 Nucleotide sequence of
Cx. quinquefasciatus
PCR amplified fragment
Notes: Total number of bases 858 bp, DNA sequence composition (A: 168; C: 248; G: 241; T: 205; N: 16)
After successful cloning of Tsf insert, it was double
digested by
Bam
HI/
Hin
dIII out of pGEM-T vector
and ligated into the expression vector pET-28a (double
digested with
Bam
HI/
Hin
dII). The ligation product
was used to transform chemically competent
BL21
(DE3)-pLysS E. coli
cells. Successfully transformed
cells were selected by growing the transformed cells
on LB agar containing Kanamycin. To confirm
insertion of Tsf in pET-28a vector, plasmid mini
preparation was prepared from transformed cells
grown successfully in the presence of Kanamycin. The
plasmid mini preparation was double digested with

