Basic HTML Version




Genomics and Applied Biology
, 2013, Vol. 4 No.1 1-7
http://gab.sophiapublisher.com
- 4 -
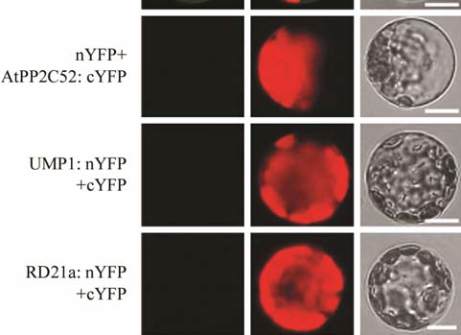
Figure 5 AtPP2C52 interacted with UMP1and RD21a in BiFC
assays in Arabidopsis protoplasts
Note: The combination of AGB1:nYFP+AtPP2C52:cYFP was
used as a positive control; The combinations of nYFP+At-
PP2C52:cYFP, UMP1:nYFP+cYFP, and RD21a:nYFP+cYFP
were used as negative controls to show that these proteins
cannot interact with half-YFP; Fluo: fluorescence from YFP;
Merge: overlap of Bright field and YFP signal; Scale
bars=25μm
2 Discussion
2.1 Expression pattern of
AtPP2C52
gene
Here we proved that P
AtPP2C52
::GUS was evident in the
whole plant in 3-week-old plants (Figure 2C), and
P
AtPP2C52
::GUS was found in most of the flower organs
(Figure 2D~F), indicating that
AtPP2C52
gene
has
a
broad expression pattern as other PP2C genes in
Arabidopsis
(
Xue et al., 2008). On the other hand, the
expression of P
AtPP2C52
::GUS was predominantly
found in vascular, root tip and apical meristem in
4-day-old seedlings (Figure 2A; Figure 2B). The
expression of P
AtPP2C52
::GUS was lower in the anther
(Figure 2D~2F) and sporangia (Figure 2G; Figure 2H).
These results suggest that the expression of
AtPP2C52
gene was in a tissue specific manner at some level.
2.2 Mutational analysis of AtPP2C52
These mutated sites of AtPP2C52 were highly
conserved in PP2C family. ABI1 is a well-known
member belonging to PP2C Group A (Sheen, 1998).
Three AtPP2C52 mutants, G99D, DGH102-104ERN
and G105D, corresponded to the ABI1 mutants,
G174D, DGH177-179KLN and G180D, respectively
(Sheen, 1998). ABI1 mutations (G174D and
DGH177-179KLN) abolished the ability of ABI1 to
block ABA-inducible transcription. G174D and
DGH177-179KLN were identified as true null
mutations of ABI1 owing to dramatic changes in
amino acids charges near to or at the PP2C active site.
G180D mutation abolished PP2C activity of ABI1.
G180D mutant was as effective as wild-type ABI1 in
abolishing the ABA-inducible transcription, suggest-
ing G180D mutant blocks ABA responses through a
dominant interfering effect without PP2C activity.
Here, G99D and DGH102-104ERN mutations of
AtPP2C52 abolished the ability of AtPP2C52 to
interact with AGB1, UMP1 and RA21a in Y2H assay
(Figure 3C, Figure 4). G105D mutation did not affect
the Y2H interaction between AtPP2C52 and UMP1
(Figure 4A). These results suggest that these residues
may essential for AtPP2C52 to bind specific targets,
such as UMP1, RA21a and AGB1. G105D may play a
different role from G99D and DGH102-104ERN.
In conclusion, physical interactions between AtPP-
2C52 with UMP1, RA21a and AGB1 were confirmed.
Mutational analysis shown that G99D, DGH102-
104ERN and G105D are essential for AtPP2C52 to
bind specific targets, such as UMP1, RA21a and
AGB1.
3 Materials and Methods
3.1 Plant materials and growth conditions
Arabidopsis Col-0 plants were used. Seeds were
surface sterilized in 70% (v/v) ethanol solution for 30s,
then sterilized in 0.25% (v/v) sodium hypochlorite
containing 1% (v/v) Tween X-100 and washed four
times in sterile distilled water. Afterward, seeds were
sowed on MS plates containing half MS basal salts
(Wako) and 1% (w/v) sucrose, pH5.7, solidified with

