Genomics and Applied Biology 2016, Vol.7, No.5, 1-8
5
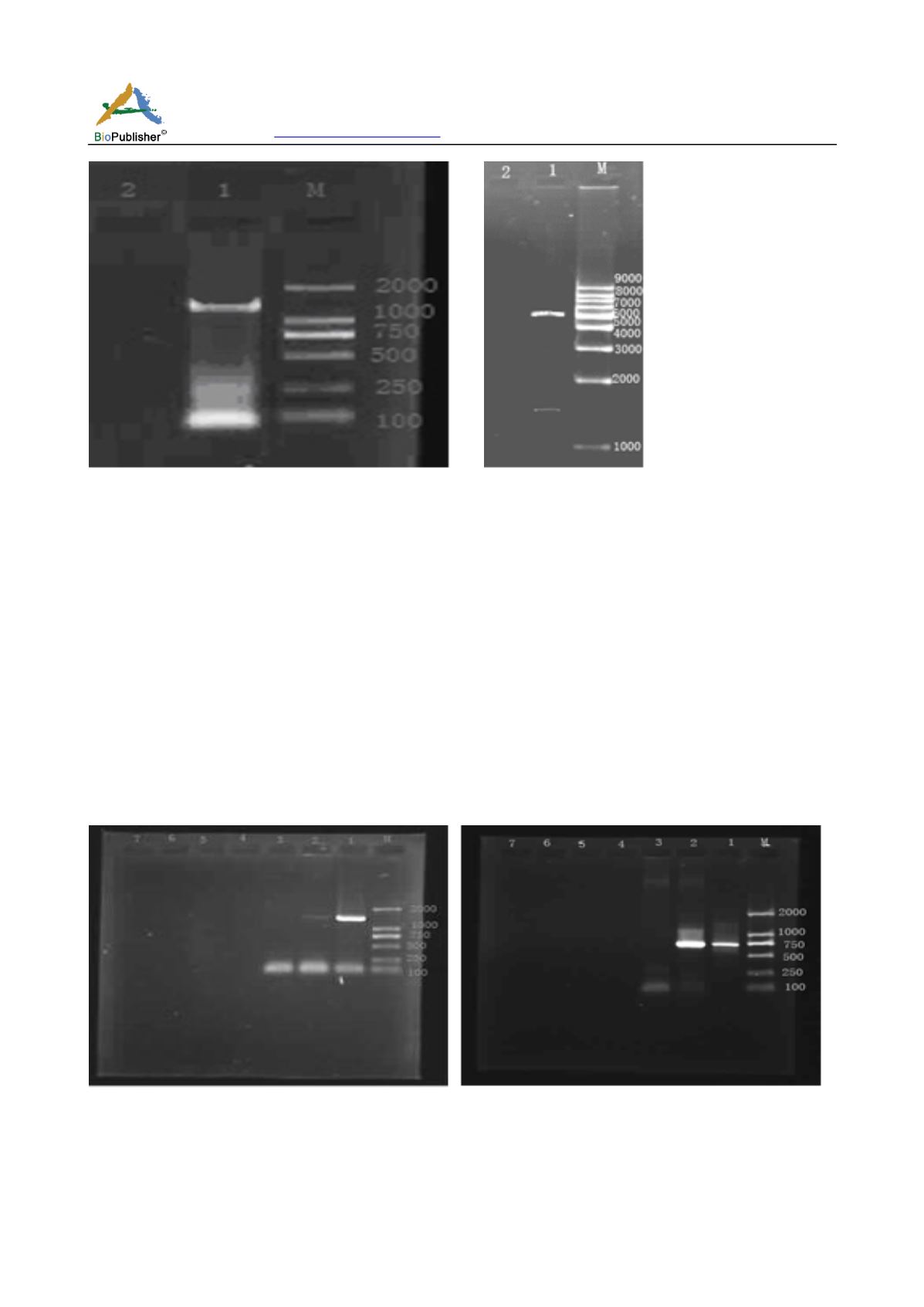
(A)
(B)
Figure 3 the detection result after cloning
Note: (A) electrophoresis of PCR product in 1% agarose gel by Ethidium bromide-staining. In lane 1, illustrate 1 401 bp product was
amplified from the recombinant plasmid using primer P1F/P2R. (B) electrophoresis of double digestion product using restriction
enzymes EcoR
Ⅰ
and Nde
Ⅰ
of 1% agarose gel by Ethidium bromide-staining. In lane 1, there are two products of 1 401 bp and 5 500
bp
Detection of HBV integration into HepG2 cells
The total DNA was extracted from HepG2 cells when the day after the HepG2 cells were transfected 4 days and
30 days, and then detected by PCR. Figure 4 (A) illustrated the whole HBV Creg DNA sequence can be amplified
by PCR in the day when the HepG2 cells were transfected 4 days (before integrated into host genome), Figure 4
(B) illustrated in the day when the HepG2 cells were transfected 30 days (after integrated into host genome), the
whole HBV Creg DNA sequence cannot be amplified by PCR using the primer P1F / P2R, but X gene which is in
the upstream of DR region can be detected using total DNA of transfected cells as templet and using primer P1F /
P1R, C gene which is in the downstream of DR region can be detected too. So the HBV Creg DNA sequence
integrates into the HepG2 genome not as a whole but upstream or downstream from DR region.
(A) (B)
Figure 4 the detection of integration function that related to HBV DR
Note: electrophoresis of PCR product in 1% agarose gel by Ethidium bromide - staining. (A) Before the HBV CregDNA integrate
into the HepG2 genome, in lane2, the whole HBV CregDNA can be detected by PCR using primer P1F / P2R and using total DNA of
transfected cells as templet, in lane1, it is positive control using pcDNA3.1 (+) - HBV Creg vector as templet, in lane3, it is negative


