基本HTML版本

Genomics and Applied Biology 2014, Vol. 5, No. 5, 1-6
http://gab.biopublisher.ca
4
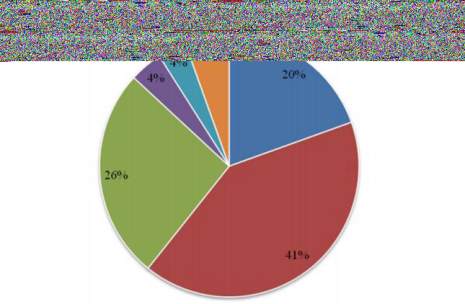
Oxidoreductases (110), Transferases (232), Hydrolases
(147), Lyases (23), Isomerases (20) and Ligases (31)
which is shown in Figure 2.
Figure 2 Enzyme Code (EC) Classification
2.3.3 Gene Ontology (GO) Classification
To functionally categorize
Phaseolus vulgaris
L.
transcript contigs, Gene Ontology (GO) terms were
assigned to each assembled transcript contigs. Out of
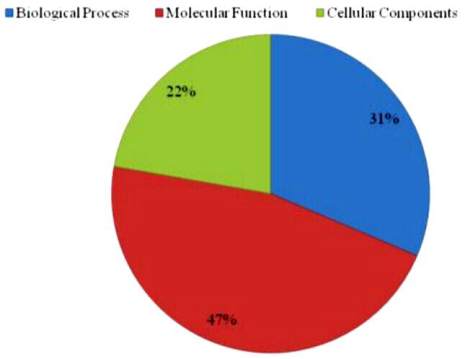
6999 transcript contigs, 3724 unigenes were grouped into
GO functional categories (http://www.geneontology.org),
which are distributed under the three main categories of
Molecular Function (1727), Biological Process (1168)
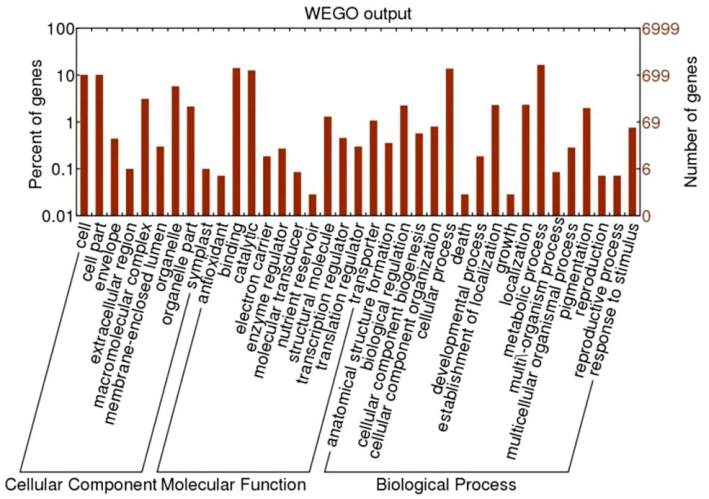
and Cellular Components (829) (Figure 3). Figure 4
which is output of WEGO tool; it shows that, Within
the Molecular Function category, genes encoding
binding proteins and proteins related to catalytic
activity were the most enriched. Proteins related to
metabolic processes and cellular processes were
enriched in the Biological Process category. With
regard to the Cellular Components category, the cell
and cell part were the most highly represented
categories.
Figure 3 Gene Ontology Result
Figure 4 WEGO Tool Result

