基本HTML版本

Computational Molecular Biology 2014, Vol. 4, No. 12, 1-8
http://cmb.biopublisher.ca
5
Figure 3 3D structure of grain heading date7 protein
Figure 4 Ramachandran plot ofaberrant panicle organisation1
protein
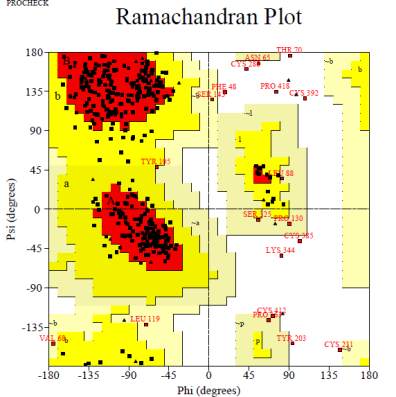
Figure 5 Ramachandran plot of dense erect panicle1 protein
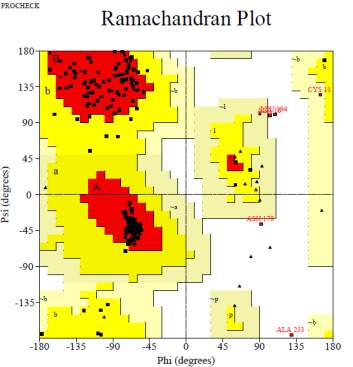
Figure 6 Ramachandran plot of grain heading date7 protein
2.4 Final primers
The final primers were selected on the basis of gc%, tm,
product size and hairpin loop sequence. The gc content
must be 40%-80%, tm must be 55
o
C-65
o
C, hairpin loop
sequence must be 0.00 and the product size must be
more depending upon the sequence length.
These primers were ordered for checking
amplification and following primers amplified
successfully whereas
Apo
1-1 promoter primer showed
negative amplification.
Four primer pairs( three for
Dep1
gene, one for
Ghd7
gene and one for
Apo1
gene were used to amplify
from high grain, HGN1 and low grain, Heera using six
different temperature regime each with six different
temperatures.
The primer pair Dep1-1 (Promoter) for
Dep1
gene
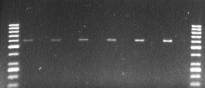
amplified band of 600bp in all the six temperatures
from high grain variety, HGN1 while fail to amplify
from Heera (Figure 7)
.
Figure 7 Gradient PCR with
Dep1
gene specific primer,
Dep1-1 (Promo 1 - HGN1 (high grain), 2- Heera( low grain)
Note: Annealing Temperatures are given on the top of gel

