基本HTML版本

Computational Molecular Biology 2014, Vol. 4, No. 12, 1-8
http://cmb.biopublisher.ca
4
2 Results and Discussion
2.1 Similarity search analysis
The
Apo1
gene showed 100% similarity with
oryza sativa
group
Apo1
for panicle organization related protein and
genomic DNAof
oryza sativa
on chromosome no.6.
Dep1
gene showed 100% similarity with genomic DNA of
japonica
group on chromosome no.9 and with
Dep1
complete cds.
Ghd7
showed 100% similarity with
indica
group cultivator
minghui63Ghd7
complete cds. The gene
prediction which was performed is given in Table 1.
Table 1 gene prediction of
Apo1, Dep1, Ghd7
high grain number genes
Gene
Gene exon type
Begin
End
Length
Fr
Ph I/ac
Do/t
Tscr
APO1
(genomic)
1.01INIT+
1.0 TERM+
166
952
869
1537
704
586
0
1
2
1
88
98
115
49
94.56
75.75
DEP1
(genomic)
1.01INIT+
1.0 TERM+
1.03PIYA+
2.03 PIYA+
2.02TERM-
2.01INIT-
2.00PROM-
3.00 PROM-
3.01 PROM-
117
288
374
589
640
824
917
1175
4183
245
356
379
484
503
681
878
1170
3296
129
69
6
6
138
144
40
6
888
2
2
1
2
1
0
0
0
0
0
85
79
39
85
_1
105
38
42
82
83
17.76
-5.66
-0.45
-1.75
-4.74
16.53
-5.56
1.06
43.66
GHD7 (genomic)
1.01
TERM+
1.02
PIYA+
2.00PROM+
2.01INIT+
110
1553
2278
2491
8181558
2317
2722
509
6
40
232
2
0
2
1
35
122
43
18
81.37
1.06
-4.76
28.37
2.2 Secondary structure prediction analysis
The secondary structure prediction of the aberrant
panicle organisation1 protein showed that it has 429
amino acids, 23.54% alpha helix, 20.28% extended
strand, 9.09% beta turn, 47.09% random coils,
whereas dense erect panicle1 protein has 426 amino
acids, 12.68% alpha helix, 9.62% extended strand,
3.76% beta turn, 73.94% random coils and grain
heading date7 protein has 257 amino acids, 17.90%
alpha helix, 13.62% extended strand, 5.45% beta turn,
63.04% random coils.
2.3 Comparative homology analysis
The final models were visualized using pymol and
discovery studio visualizer. The backbone
confirmation of all the proteins were analyzed which
showed that
Apo1
protein lies in 84.9% of allowed
region and 0.9% of disallowed region,
Dep1
protein
lies in 80.6% of allowed region and 2.6% of
disallowed region,
Ghd7
lies in 85.4% of allowed
region and of disallowed region. The homology
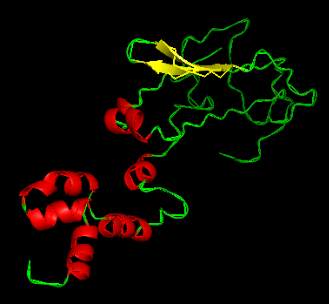
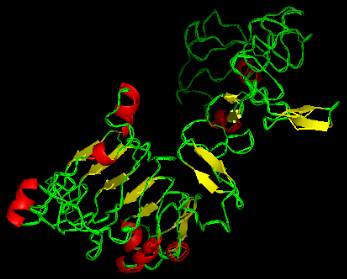
models are given in Figure 1-Figure 3 and the
backbone confirmations of all the tertiary structures
are given in Figure 4-Figure 6.
Figure 1 3D structure of aberrant panicle organisation1 protein
Figure 2 3D structure of dense erect panicle1 protein

