基本HTML版本

Computational Molecular Biology 2014, Vol. 4, No. 10, 1-17
http://cmb.biopublisher.ca
5
Table 1 Percentage of overlap PhastCons conservation regions for all H3K4me co-localized peaks in TPRs
OR cutoff
pC cutoff
me1me2
me1me2me3
me1me3
me2me3
Single
0.1
0.2
30%
49%
49%
45%
44%
0.1
0.6
24%
41%
42%
37%
37%
0.5
0.2
30%
48%
48%
45%
44%
0.5
0.6
24%
40%
41%
37%
37%
1.0
0.2
32%
57%
45%
48%
44%
1.0
0.6
25%
49%
40%
41%
36%
Note: OR cutoff is Overlap rate cutoff. pC cutoff is phastCons score cutoff.
Because co-localized peaks except me1me2 were
more conserved than single-localized peaks, we used
known protein–protein interaction (PPI) data to
further confirm the finding. Proteins having multiple
partners were considered to be conserved and
functionally important. PPI data were collected from a
manually curated PPI database: HPRD
. From Table 2, we observed that
the co-localized peaks had overall more partners than
single-localized peaks (
p
< 0.05). Though the average
partner number for me1me3 (11.99) was the largest, it
was strange that me1me3 was not significant against
the control group (
p
= 0.07). We found a protein
named with CREBBP with degree of 199 in PPI
network, which biased the average degree of me1me3.
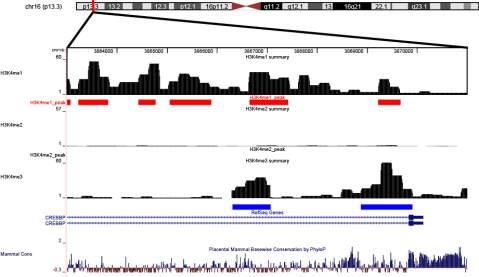
The
CREBBP
gene was visualized in the UCSC
Genome Browser
, which was
shown in Figure 2. In Table 1, H3K4me triplet type
seemed the most conserved. But in Table 2, the partner
number for triplet type was lower than those of me1me3
and me1me3, which may be caused by the least gene
number for the triplet type (gene number : 536).
Figure 2 The
CREBBP
gene with surrounding context is displayed as custom tracks on the UCSC genome browser. Red frame
indicates the [-1k, 2k] around TSS. The red rectangular track indicates the peaks detected by Cisgenome

