Cotton Genomics and Genetics 2016, Vol.7, No.2, 1-23
4
sequences deposited at miRBase has identified several conserved miRNAs (Supplementary Table 1). For effective
comparison of results, only miRNAs that have shown more than two fold change in RPKM either in up or down
regulation were considered and discussed hereunder.
Table 2 Statistics of differentially expressed small RNA sequence reads in two different cotton accessions under two different water regimes
Note: * Total number of distinct reads found in each comparison;
#
number of reads that match with
G. raimondii
genome
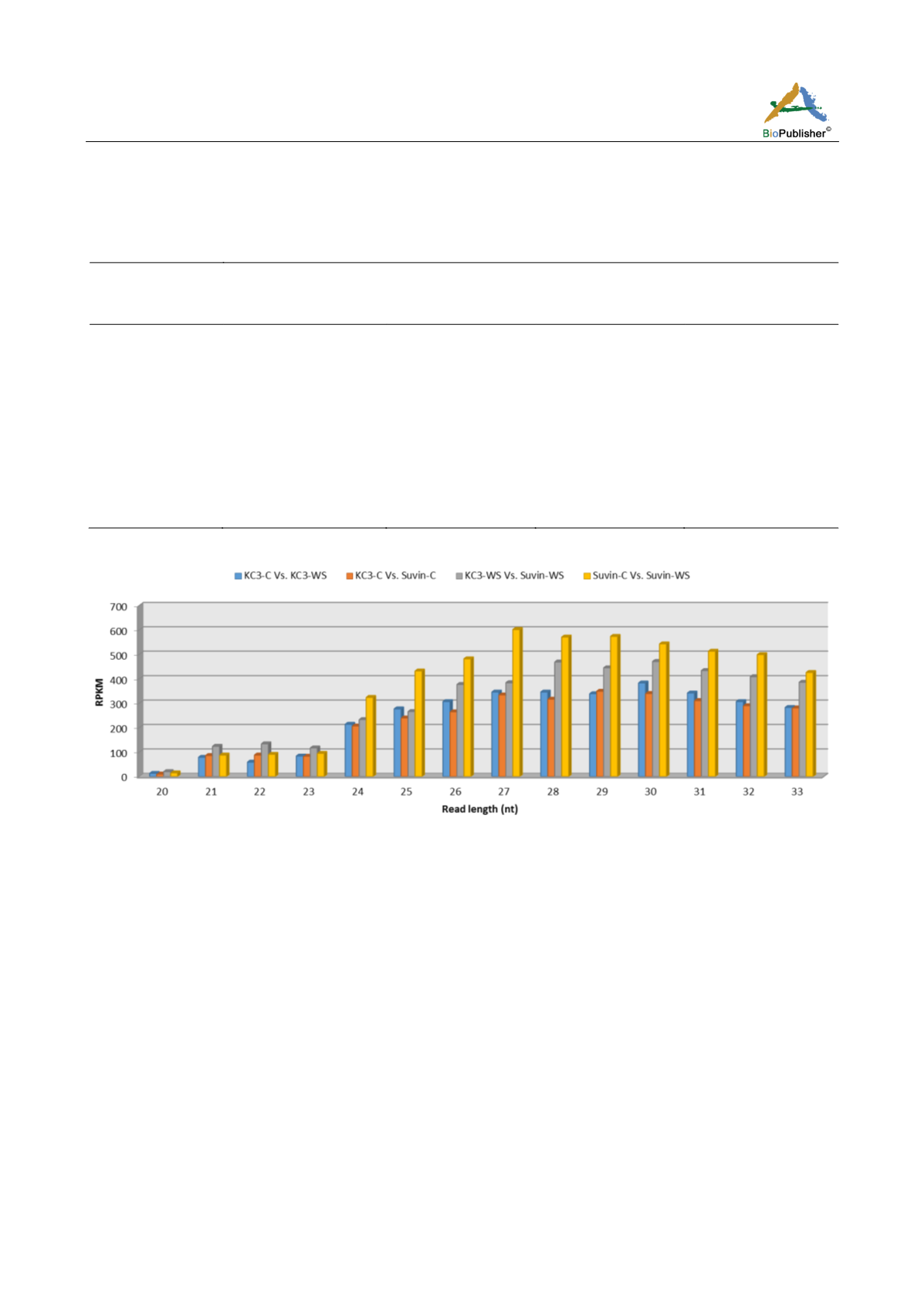
Figure 1 Sequence length distribution of small RNAs that were differentially expressed in two different cotton accessions under two
different water regimes
Seventy one conserved miRNAs were found as differentially expressed between the miRNA libraries prepared
from KC3-C and KC3-WS (Supplementary Table 1). Among them, 35 (belonging to 14 miRNA families) were up
regulated and 36 (belonging to 21 miRNA families) were down regulated. Interestingly, members of three of the
miRNA families (
viz
., miR166, miR167 and miR399) were both up and down regulated (Supplementary Table 1).
Among the up-regulated miRNAs, miR398 family was highly induced under water stress in KC3 (38.880 folds),
followed by miR397 (13.256 folds) and miR10 (8.242 folds). On the other hand, expression of miR166 was found
to be highly down regulated under water stress in KC3 (54.840 folds), followed by miR479 (46.919 folds) and
miR2111 (29.654) (Supplementary Table 1). Interestingly, several miRNA families were found to exhibit more
than 10 fold change in their expression level in KC3-WS (for example, miR397 and miR398 were more than 10
fold up regulated and miR160, miR164, miR166, miR390, miR399, miR479, miR2111 and miR2796 were more
than 10 fold down regulated; Supplementary Table 1). In contrast, some of the miRNA family had lowest fold
change (miR535 was in 2.031 fold up regulation and miR160 was in 2.060 fold down regulation).
When comparing the miRNAs that were differentially expressed between the miRNA libraries prepared from
drought tolerant, KC3 and drought susceptible, Suvin under water stress in the natural field conditions, totally 142
conserved miRNAs that belongs to 29 conserved families were noticed (Supplementary Table 1). It was found
Description
Differentially expressed reads in
KC3-C
Vs.
KC3-WS
KC3-C
Vs.
Suvin-C
KC3-WS
Vs.
Suvin-WS
Suvin-C
Vs.
Suvin-WS
Total number*
5138
5341
7494
8469
Match genome
#
789 (15.35%)
759 (14.21%)
932 (12.43%)
1250 (14.75%)
rRNA
4
6
6
7
snRNA
1
1
5
3
snoRNA
11
10
15
10
tRNA
20
19
28
25
tmRNA
0
0
0
1


