Cancer Genetics and Epigenetics 2016, Vol.4, No.2, 1-9
4
- cytokine receptor interaction, retinol metabolism, drug metabolism, complement and coagulation cascades,
metabolism of xenobiotics by cytochrome P450, steroid hormone biosynthesis and ECM-receptor interaction
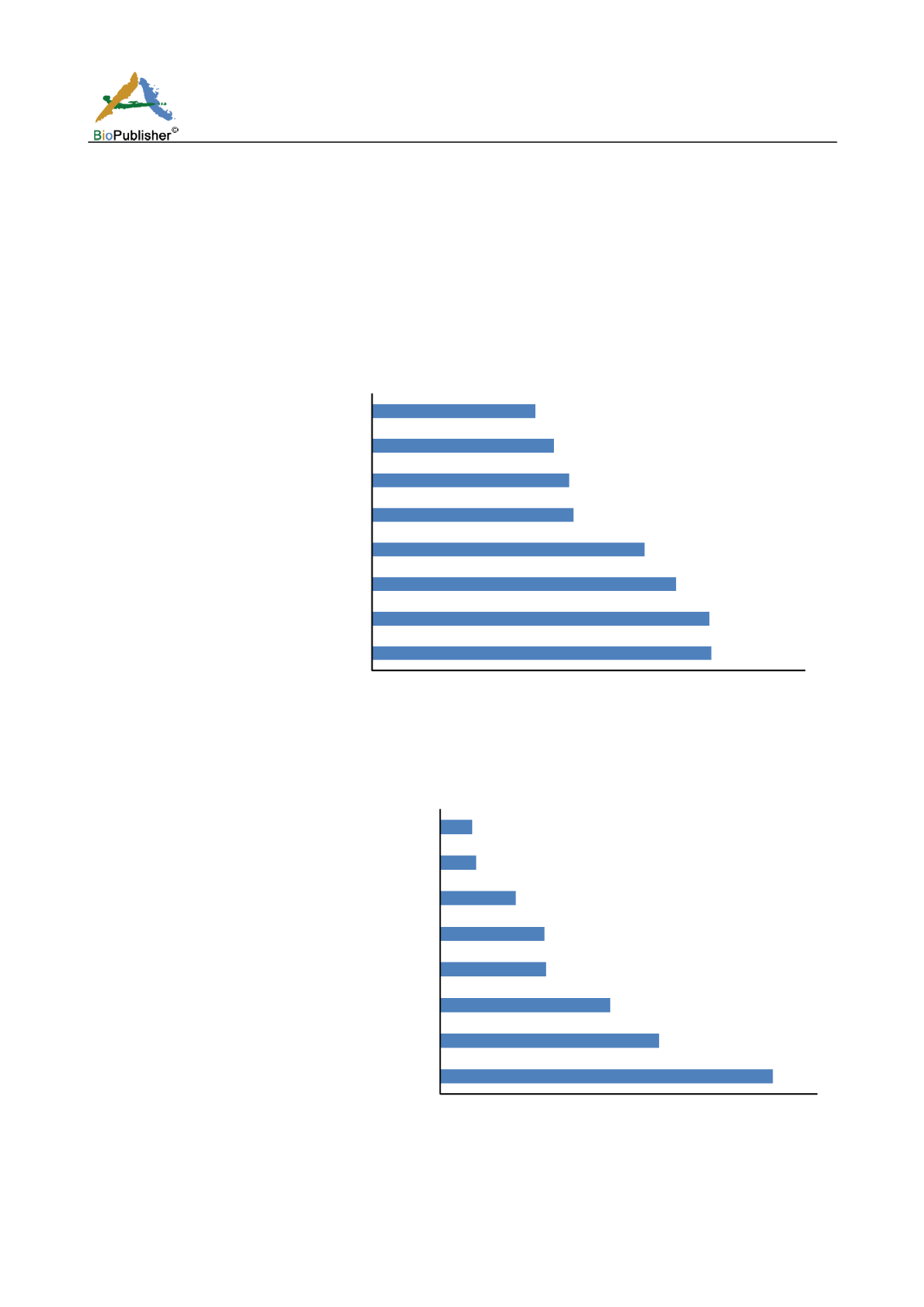
(Figure 1). A graph in figure 1 illustrated the results of GO functional analysis; we show only the most significant
eight GO terms, B graph was the results of KEGG pathway analysis, showing all eight significant pathways. In
order to obtain information on the interaction between genes in these pathways, we download the XML files of
these eight pathways from the KEGG website, in which including information on gene interaction. The
information of these eight pathways will be used for further analysis.
Figure 1 Gene Ontology function and KEGG pathway enrichment analysis.
A. Gene Ontology function enrichment analysis, only the
most significant eight go terms were shown in this figure. B. KEGG pathway enrichment analysis
0
2
4
6
8
10
12
14
16
hsa04080:Neuroactive ligand-receptor interaction
hsa04060:Cytokine-cytokine receptor interaction
hsa00830:Retinol metabolism
hsa00982:Drug metabolism
hsa04610:Complement and coagulation cascades
hsa00980:Metabolism of xenobiotics by cytochrome P450
hsa00140:Steroid hormone biosynthesis
hsa04512:ECM-receptor interaction
-log(p)
KEGG pathways
0
5
10
15
20
25
GO:0007155~cell adhesion
GO:0022610~biological adhesion
GO:0007267~cell-cell signaling
GO:0007610~behavior
GO:0044057~regulation of system process
GO:0006811~ion transport
GO:0007156~homophilic cell adhesion
GO:0016337~cell-cell adhesion
-log(p)
Biological processes
A
B


