Basic HTML Version

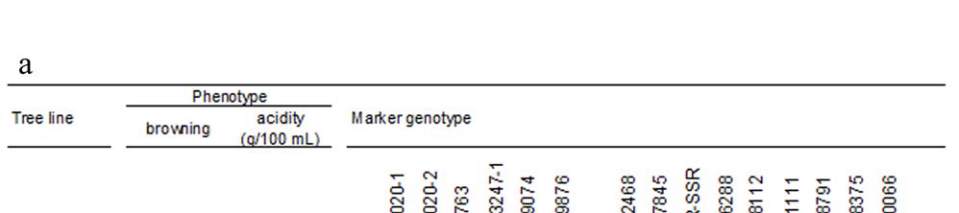

Tree Genetics and Molecular Breeding 2014, Vol.4, No.2, 1
-
10
http://tgmb.biopublisher.ca
7
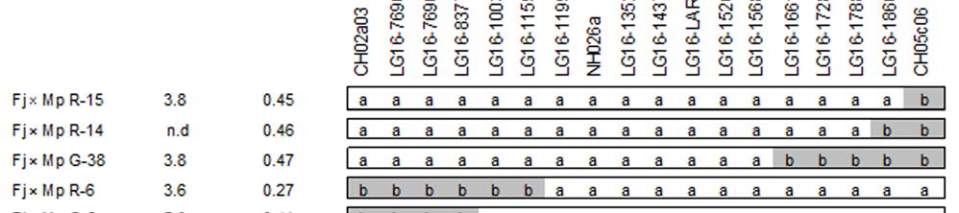
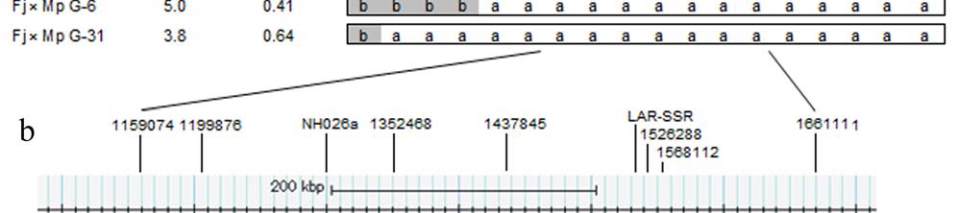
Figure 5 Genetic and physical mapping of the QTLs for fruit juice browning and fruit acidity on apple chromosome 16
Note: a: Genotyping of recombinants around the QTL region.The
a
and
b
represent allels linked to the high browning/low acidity and
low browning/ high acidity phenotype, respectively. b: QTL region defined by the two flanking markers, LG16-1159074 and
LG16-1661111, the physical locations of each SSR markers and candidate genes within the QTL region of ‘Golden Delicious’
genome sequence (a screenshot from the Genome Browser of
Malus
×
domestica
at Genome Database for Rosaceae (GDR) BLAST
server (http://www.rosaceae.org))
with degree of fruit juice browning, as the lower pH of
fruit juice tends to be less susceptible to
PPO-catalyzed browning (Sun-Waterhouse et al.,
2011). We observed a similar trend in the Fj × Mp
population, with fruit juice of high-acid fruit progeny
tending to exhibit less browning than that from
offspring having low acidity (Figure 3 and Figure 4).
Thus, the fruit acidity QTL (the
Ma
locus, ALMT)
may be associated with the degree of fruit juice
browning through the regulation of pH and PPO
activity. This trend, however, does not seems to be
true for all apple cultivars, as there are some cultivars
that bear low-acidity, low-browning fruit, such as
‘Tsugaru’ (‘Golden Delicious’ × ‘Jonathan’) and
‘Michinoku’ (‘Kitakami’ × ‘Tsugaru’).
Second, LAR located within the QTL region is a
possible candidate for determining polyphenol content

