Basic HTML Version



Rice Genomics and Genetics 2013, Vol. 4, No. 5, 22-27
http://rgg.biopublisher.ca
26
inference is in agreement with other workers who used
the same AMMI model for their analysis. Vijayakumar
et al., (2001) used bi-plot (AMMI) assay and
identified two hybrids viz., IR58025A/Swarna and
IR58025A/IR21567 as having general adaptability at
all locations. They also reported that hybrids ORI 161
and IR58025N1R54742 were identified as specifically
adapted to favourable locations. Several authors have
used AMMI to evaluate multi-environment
experiments to distinguish the effects of the genotype
and the environment and then assess the G × E
interaction in a reduced dimensional space with
minimum error (Kandus et. al., 2010).
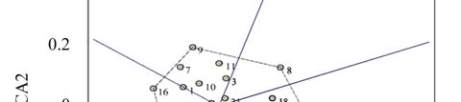
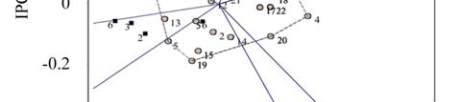
Figure 2 Projection of 22 selected genotypes on the first two
principal components of GEI effect
Note: 1~12: BIL-5, BIL-6, BIL-48, BIL-50, BIL-57, BIL-60,
BIL-73, BIL-77, BIL-83, BIL-90, BIL-93, BIL-118, BIL-142,
BIL-149, BIL-150, BIL-163, BIL-174, BIL-183, BIL-188,
Swarna, Prasanna, MGD-101;
2 Material and Methods
The breeding material for this study was inter-varietal
backcross inbred lines (BILs) of Swarna × WAB 450
developed at Barwale Foundation, Hyderabad. Swarna
is a mega rice variety from India whereas WAB 450 is
an inter-specific derivative of NERICA lines from
Africa. Multi-location yield trials of 19 superior BILs
(BC1F8) selected for earliness, productivity, reaction
to blast diseases and grain size were conducted at 3
locations-Mugad, Gangavati and Sirsi during
kharif
and summer seasons in 2011 and 2012, respectively.
The selected BILs including recurrent parent (Swarna)
and 2 released local varieties (Prasanna and MGD 101)
were evaluated for yield in six environments of which
3 environments were in
Kharif
and 3 in summer
season, respectively. All trials were laid out in
randomized block design with 2 replications. In each
trial, 25~30 days old seedlings were transplanted in 20
cm × 15 cm spacing and one seedling per hill. Normal
cultural practices and plant protection measures were
followed in each trial according to package of practice
developed by University of Agricultural Sciences
Dharwad (UAS Dharwad, 2009). In all trials, data
were recorded on net plot for grain yield. Grain yield
data of the six environments was analyzed for G × E
interaction using AMMI model (Zobel et al., 1988) to
identify genotypes adapted to specific environments.
3 Conclusion
Six genotypes [genotypes 3, 21, 11, 2, 12 and 14]
were found to be indicating small effect of GE
interaction in the present study; three out of these six
genotypes (2, 12 and 14) had mean yield level
between 5000 and 5650 kg/ha reflecting greater
breeding advances and hence they are recommended
for all environments. Genotype 18 with high mean
yield level and positive IPCA 1 is recommended for
favourable environment whereas genotypes 5 and 13
with high mean yield level and negative IPCA 1 are
recommended for unfavourable environment. It can be
concluded based on the values of mean and ICPA 1 for
all 22 genotypes evaluated in this study that 6
genotypes were found suitable for all environments, 6
genotypes for favourable environments while 10
genotypes were identified as suitable for unfavorable
environments.
References
Becker H.C. and J. Leon, 1988, Stability analysis in Plant
Breeding, Plant Breeding, 101: 1-23
http://dx.doi.org/10.1111/j.1439-0523.1988.tb00261.x
Crossa H.M., 1990, Statistical analyses of multi-location trials,
Advances in Agronomy, 44: 55-85
http://dx.doi.org/10.1016/S0065-2113(08)60818-4
Das S., Misra R.C., Patnaik M.C. and Das S.R., 2010, G × E
interaction, adaptability and yield stability of mid-early
rice genotypes, Indian J. Agric. Res., 44(2): 104-111
Dabholkar A.R., 1992, Elements of Bio Metrical Genetics,
Concept publishing Company. New Delhi, pp.379-421

