Basic HTML Version


Rice Genomics and Genetics 2013, Vol. 4, No. 5, 22-27
http://rgg.biopublisher.ca
25
Continuing Table 2
A
B
C
D
E
F
G
H
I
J
18
BIL-183
4702.75 6579.82 6394.17 5400.00 6628.21 5538.00 5873.82 -6.63
19
BIL-188
7274.29 4777.60 4916.98 6150.00 6445.51 4705.50 5711.65 -6.11
20
Swarna
5387.15 5235.46 5858.71 4350.00 4852.57 5513.00 5199.48 -12.17
21
Prasanna
4059.77 5283.23 5899.79 5600.00 4178.85 3268.00 4714.94 -13.90
22
MGD 101
4889.48 6326.34 6246.30 5100.00 6467.95 5228.00 5709.68 -4.57
SITE MEAN 4870.86 5329.51 5292.55 6095.45 5781.29 4540.93
Note: A: Genotype Code No; B: Genotype Name; C:
Kharif
2011 Mugad; D:
Summer
2011 Mugad; E:
Summer
2012 Mugad; F:
Summer
2011 Gangavati; G:
Kharif
2012 Gangavati; H:
Kharif
2012 Sirsi; I: Treatment Mean; J: PCA 1
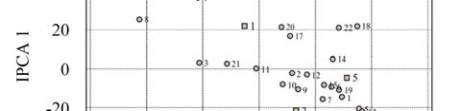
Figures 1 and Figure 2 presented bi-plot assays of the
AMMI results. Figure 1 showed the main effects
[genotype means and environment means] on the
abscissa (x-axis),
and the ordinate (y-axis)
representing the first PCA. Both main effects and
interaction component are shown clearly in the figure.
Favourable environment is represented by positive
PCA and unfavourable environment was represented
by negative PCA. Result showed that six genotypes
[genotypes 3, 21, 11, 2, 12 and 14 (Figure 1)] have
PCA near to zero indicating small effect of GE
interaction, these genotypes have differences only in
main (additive) effect. Three out of these 6 genotypes
(2, 12 and 14) have mean yield level between 5000
and 5650 kg/ha with genotype no 14 being the highest
(Figure 1). Genotype 18 recorded very high yields and
high PCA score in favourable environment whereas
genotypes 5 and 13 recorded high mean yield level
and had high PCA scores in unfavourable environment
(Figure 1). Whereas genotypes 18, 5 and 13 had
differences only in interaction effect, genotypes 3, 21,
11, 2, 12 and 14 have differences only in main effect.
Figure 2 is the projections of the genotypes on the
environmental vector. Result also indicated that only
one environmental E5 (Gangavati,
Kharif
2012) had
PCA near to zero. Whereas environmental mean E1
(Mugad
Kharif
2011) and E4 (Gangavati summer
2011) had positive PCA the environments E2 (Mugad
summer 2011), E3 (Mugad summer 2012), E5
(Gangavati
Kharif
2012) and E6 (Sirsi
Kharif
2012)
had negative PCA (Figure 1). Interactions of
environments were highly varied; whereas E5 has low
interaction, E4 and E6 were highly interactive. Figure
2 presents the spatial pattern of the first two PCA axes
Figure 1 Bi-plot graph for 22 selected genotypes mean and
interaction principal component-1 (IPCA1)
Note: 1~12: BIL-5, BIL-6, BIL-48, BIL-50, BIL-57, BIL-60,
BIL-73, BIL-77, BIL-83, BIL-90, BIL-93, BIL-118, BIL-142,
BIL-149, BIL-150, BIL-163, BIL-174, BIL-183, BIL-188,
Swarna, Prasanna, MGD-101;
of the interaction effect corresponding to the
genotypes. These bi-plots help in visual interpretation
of the GE patterns and identify genotypes or locations
that exhibit low, medium or high levels of interaction
effects (Vijayakumar et al., 2001). We can infer from
the result of this study that genotypes 11, 2, 12 and 14
had mean yield levels higher than that of local check
(Prasanna-genotype 21), and were less influenced by
the GEl effect and when compared to check were
more widely adaptable. The bi-plot showed that
genotypes 12 and 21 were more stable as they were
located near the origin (Figure 2). It can be inferred
from the result of this investigation that genotypes 5, 6,
2, 14, 15 and 19 are specifically adapted to
environment E6 while genotype 13 is adaptable to
three environments [E2, E3 and E6 (Figure 2)]. This

