Basic HTML Version


Molecular Plant Breeding 2010, Vol.1 No.4
http://mpb.sophiapublisher.com
Page 2 of 5
Wang et al. (2007) found two Linked markers
CINAU17
-
1086
and
CINAU18
-
723
located in 0.58 ~
0.70 and 0.00~0.45 section on 6VS respectively, when
checking with 11 RGA (Resistance Gene Analogy)
and 17 pairs of STS primers; Wang et al. (2007)
developed a RAPD primers (
OPK08
910
) and three
AFLP primers which can be used to select the
Pm21
gene from
Haynaldia villosa
; Wei et al. (2009)
developed to a EST-PCR marker (
XCAU127
) to
efficient distinguish the
Pm12
and
Pm21
. As the
RAPD markers were low stability and poor
reproducibility, and the ARLP markers were technical
cumbersome, expensive and difficult to automate,
both were limited in plant breeding. Until now, the
molecular markers used in marker-assisted selection
for the
Pm21
gene mainly focused on
SCAR1265
(Liu
et al., 1999),
SCAR1400
(Liu et al., 1999), STS
marker
CINAU15
902
(Cao et al., 2006),
CINAU16
1650
(Chen et al., 2006),
CINAU17
1086
(Wang et al., 2007),
CINAU18
723
(Wang et al., 2007) and EST-PCR marker
XCAU127
(Wei et al., 2009).
In this study, 258 SSR or STS markers located on
wheat homoeologous group VI, were used to identify
the linked molecular markers for the gene
Pm21
, and
the obtained markers will enrich the selection
strategies for this gene and enhance the selection
accuracy in breeding.
1 Results
1.1 Identification of wheat powdery mildew resista-
nce
The resistant parent CB033 showed immune to
mildew when the susceptible control Huixianhong
fully infected. For the F
2
segregation population of
CB033 / Huixianhong (213 individuals), the resistant
plants were 153 individuals, and the susceptible plants
were 60 individuals. The difference of chi-square text
was not significant (χ2 = 1.401 <χ2
0.05
, P> 0.05), the
separation ratio of resistant and susceptible plants was
3:1 (Table 1).
1.2 Screening of polymorphic primers
258 SSR or STS markers located on wheat homoeolo-
gous group VI, were used to the initial screening for
the parents CB033 and Huixianhong, 102 markers in
the parents revealed polymorphism. Then the 102
markers were used to the preferred small groups
analysis, and two EST-SSR markers
Xcfe164, Xedm129
and one STS marker
Xcinau188
amplified polymorphic
bands in the preferred small groups.
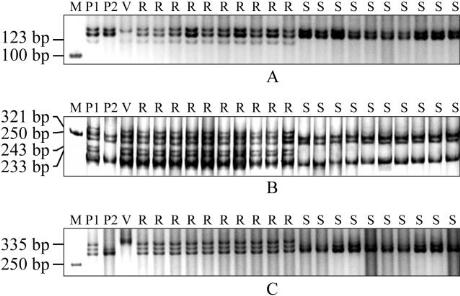
Xcfe164
could
amplify a band about 123 bp size in the 10 typical
resistant individuals, and could not amplify the band
in the 10 typical susceptible individuals (Figure 1A);
Xedm129
could amplify there bands respectively
about 233 bp, 243 bp and 321 bp size in the 10 typical
resistant individuals, and could not amplify the three
bands in the 10 typical susceptible individuals (Figure
1B);
Xcinau188
could amplify a band about 335bp
size in the 10 typical resistant individuals, and could
not amplify the band in the 10 typical susceptible
individuals (Figure 1C).
1.3 Linkage Analysis
To further test whether these three markers linked
with the gene
Pm21
, F
2
segregating population was
used to linkage analysis. The results show that, the
three markers are all closely linked with the gene
Pm21
, the resistant and susceptible genotypes in the
F
2
segregating population are consistent with the
expected separation ratio (3:1) (Table 1); the linkage
distance between
Pm21
and molecular markers of
Xcfe164
,
Xedm129
and
Xcinau188
were 3.75 cM,
1.26 cM and 0.98 cM, respectively (Figure 2). Part F
2
plants of the PCR results (Figure 3).
Figure 1 Amplification products by
Xcfe164, Xedm129
and
Xcinau188
in resistant parent CB033 (P1), susceptible parent
Hui xianhong (P2), 10 typical resistant individuals and 10
typical susceptible individuals of F
2
.
Note: A:
Xcfe164
; B:
Xedm129
; C:
Xcinau188
; R: Resistant
plants; S: Suspectible plants; P1: CB033; P2: Hui xianhong; V:
Haynaldia villosa
; M: DL2000 marker

