Basic HTML Version



Genomics and Applied Biology, 2010, Vol.1 No.2
http://gab.sophiapublisher.com
- 18 -
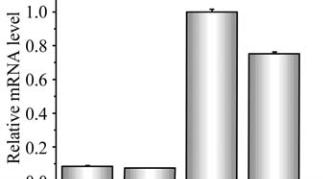
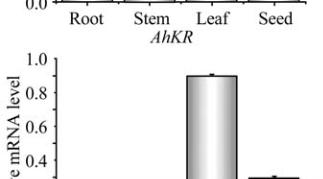
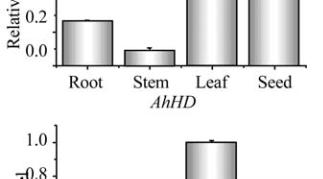
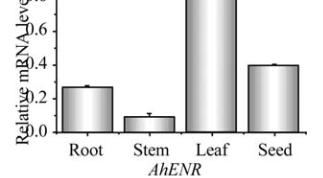
Figure 8 Expression analysis of
AhKR
,
AhHD
and
AhENR
genes in seed at different developmental stages
that the three genes may have different biochemical
functions during vegetative growth and seed
development. To improve peanut oil quality should
coordinate the expression of the three enzymes
involved in the fatty acid synthesis pathway. This
work may provide a basis for elucidating the
molecular mechanism of fatty acid synthesis and
provide candidate genes for modifying oil quality via
transgenic plants.
3 Materials and methods
3.1 Plant materials
Peanut seeds (
Arachis hypogaea
L. cultivar Huayu19)
were sown in sand and soil mixture (1:1), grown in a
growth chamber under a 16~8 h light-dark cycle at
26°C and 22°C, respectively. Three kinds of
12
-
day-old tissues including root, stem and leaf were
collected as experimental materials for quantitative
real-time RT-PCR analysis. In addition, the immature
peanut seeds from 25 to 60 days after pegging (DAP)
were also collected for expression analysis.
3.2 Nucleic acid manipulation
Total RNA was extracted from samples using the
RNeasy Mini Kit (Qiagen) according to the manufa-
cturer’s instructions. The RNA samples were used for
real-time RT-PCR after RQ1 RNase-free DNaseI
(Promega, Wisconsin, USA) treatment to remove
genomic DNA. The first-strand cDNA was synthesized
with RT-PCR kit (Promega, Wisconsin, USA) using
500 ng of total RNA according to the manufacturer’s
instructions. Controls received water instead of
reverse transcriptase to assess any contamination from
genomic DNA as described by Zhou et al. (2007).
3.3 Full-length cDNA sequence isolation
PCR was performed with the LA PCR system (Takara)
using 2.5 μL of 10×PCR buffer with MgCl
2
, 1 μL of
10 μM each primer, 4.0 μL of 10 mM dNTPs, 1 μL
cDNA samples and 0.5 μL LA
Taq
™ DNA
polymerase, and 15 μL double distilled water. The
PCR products were run on 1% agarose gel and
purified with Gel Extraction Kit (Takara) according to
the manufacturer’s protocol. The purified products
were then cloned into the pMD18
-
T Easy vector
(Takara) and sequenced (Shangon, Shanghai) (Table 2).
3.4 Multiple sequence alignment and phylogenetic
analysis
Physicochemical properties of the deduced protein
were predicted by Protparam (http://www.expasy.ch/
tools/protparam.html). The putative subcellular localiza-
tions of the candidate proteins were estimated by
TargetP (http://www. cbs.dtu.dk/services/TargetP/)
and Predotar (http://urgi.versailles.inra.fr/ predotar/
predotar.html). The potential N-terminal presequence
cleavage site was predicted by ChloroP (http://www.
cbs.dtu.dk/services/ChloroP/). Amino acid sequences
were aligned using ClustalX program with the
implanted BioEdit (Thompson et al., 1994). The
neighbor-joining (NJ) method in MEGA4 (Tamura et
al., 2007) was used to construct the phylogenetic tree.
Bootstrap with 1000 replicates was used to establish
the confidence limit of the tree branches. Default
program parameters were used.

