Molecular Plant Breeding 2015, Vol.6, No.16, 1
-
13
2
morpho-physio-chemical characters, but this method
is slow, unrealistic and identification based on
morphological traits is prone to environmental
variations. At the same time, a low level of genetic
diversity exists within and among cultivated tobacco
types (Ren and Timko, 2001). Cultivars that are
closely related or less diverse cannot be easily
distinguished by morphological indices (Degani et al.,
1998). Accurate identification of cultivars has become
increasingly important in realm of Plant Breeders
Rights and cultivar registration. Hence, the molecular
marker-based diversity analysis among the cultivars
will provide the precise data for cultivar identification.
The frequency of microsatellites in plant genomes is
low compared to animal genomes (Maguire et al.,
2000; Squirrell et al., 2003) and this feature makes the
isolation of microsatellites, a technically demanding
task. With an objective of developing more SSR
markers, the present study helped in the identification
of microsatellite motifs using microsatellite enriched
library, developed 70 SSR markers and demonstrated
their use in understanding of genetic diversity among
different types of tobacco. In addition to this,
transferability of these markers across different
species of
Nicotiana
was tested and a minimal set of
markers that could differentiate various species of
Nicotiana
was identified.
2 Results
2.1 Identification of microsatellites loci from the
tobacco enriched library
Screening of microsatellite-enriched library of
N.
tabacum
cv. Jayasree resulted in the identification of
1152 putative recombinants, which were then sequenced.
Good quality sequences were obtained for 946 clones
and all sequences (100%) contained microsatellite
motifs. However, only 224 sequences were considered
for designing primers. Considering the 224 clones
possessing class I and class II SSRs as described by
Mc Couch et al., 2002, AG/TC repeats were found to
be abundant (56 clones, 25.3%) followed by CT/GA
(38 clones, 17.2%), AC/TG (25 clones, 11.3 %),
GT/CA (20 clones, 9.0%) and rest of the clones
contained AG and CT compound repeats. The repeat
length was highly variable at these loci with maximum
for TbM13 with (GA)
29
and TbM22 with (AGA)
24
.
These sequences were submitted to NCBI Genbank
and details of the sequence with accession numbers
were tabulated (Table 2).Seventy motifs containing
class I SSRs were targeted for marker development, of
which, thirty nine (55.8%) were perfect repeats
[(TC/AG)
n
, (GT/CA)
n
, (GA/CT)
n
and (AC/TG)
n,
], five
(7.1%) were compound repeats and twenty six (37.1%)
were interrupted repeats. The homology searches with
the primers of these markers against the primers of
markers reported earlier by Bindler et al., 2011; Tong
et al., 2012 showed no significant homology.
2.2 Diversity among Flue Cured Virginia (FCV)
tobacco
All the 70 SSR markers had shown clear and robust
amplification with expected product size. Further,
these markers were used in the diversity analysis of 24
FCV varieties of
N. tabaccum
. The pair-wise
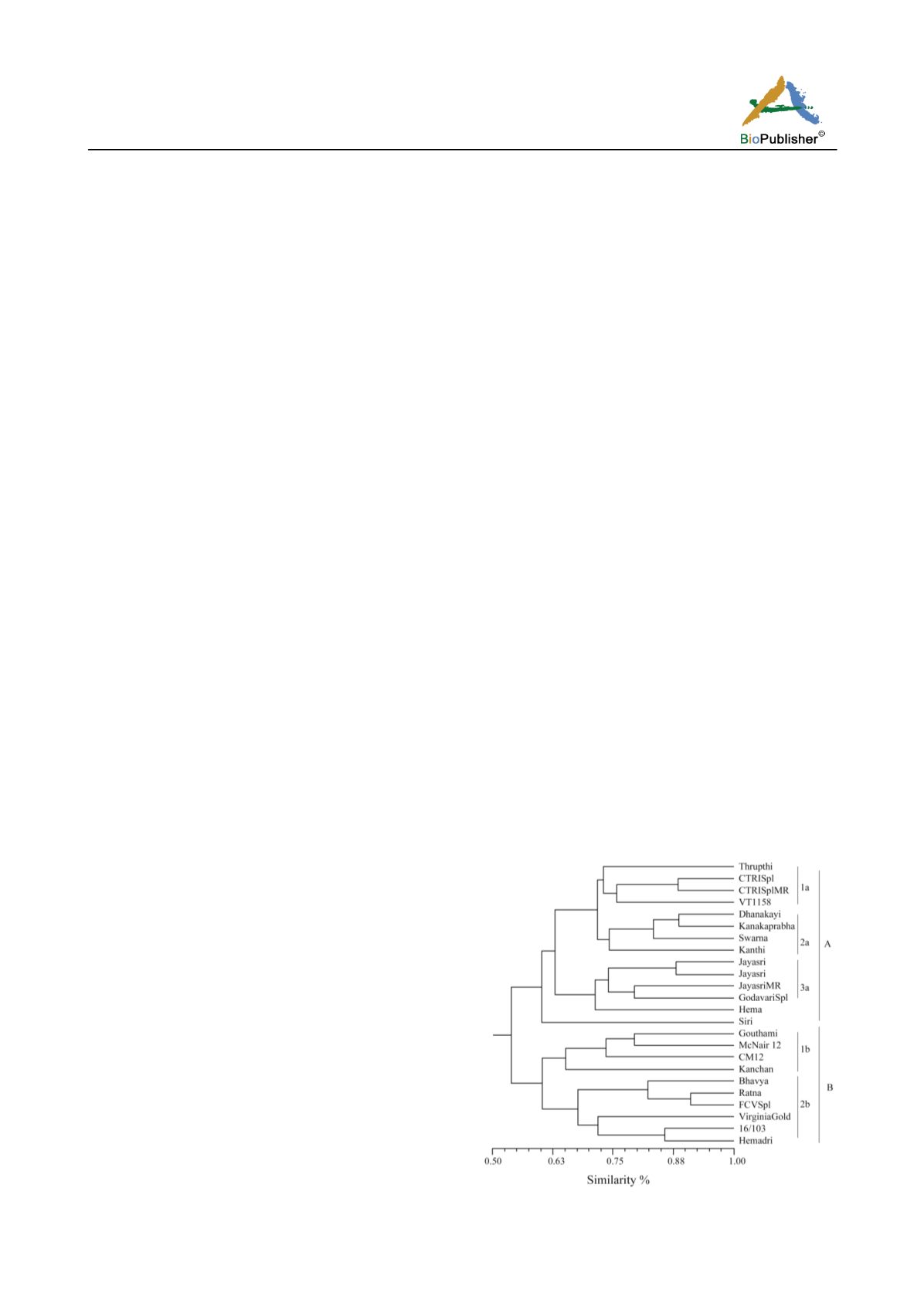
similarity measures among the tested varieties ranged
from 0.54 to 0.91 revealing a broad genetic base
(Figure 1). The varieties were mainly grouped into
two main clusters. Cluster-A consisted of varieties
developed indigenously by different breeding methods,
while Cluster-B consisted of exotic introductions and
their derivatives. The mean genetic similarity among
the indigenously developed cultivars was 0.72.
2.3 Characterization of different types of tobacco:
The microsatellite marker analysis of different types
of Indian tobaccos revealed that the varieties
belonging to the same category were grouped together.
There were different clusters for each tobacco type
and the clustering pattern supports the traditional
classification, except for the grouping of GC-2
(chewing type) and Dharla (Hookah type) in the same
Figure 1 Clustering of tobacco varieties using SSR markers


