Basic HTML Version
Computational Molecular Biology
2
In the present study, the three dimensional structures
of SoxY, SoxZ and 5’-nucleotidase C-terminal domain
of SoxB from
A.vino
obtained by homology modeling
have been described. Molecular docking simulations
have been performed in order to find out the possible
modes of binding of these proteins. Binding sites of
SoxY, SoxZ and SoxB have been predicted and
analyzed. These studies provide a detailed structural
view of the acceptable residue level interaction of
these proteins in the global sulfur oxidation
reaction cycle.
1 Results
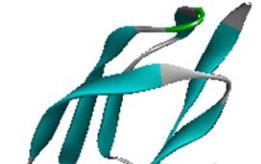
1.1 Description of the structure of SoxY
The modelled structure of SoxY is a 111 amino acid
residue long protein. The predicted structure is very
similar to the structure of the Sulfur Carrier Protein
SoxY from
Chlorobium Limicola F Thiosulfatophilum
(PDB code: 2NNC A chain for SoxY). The most of the
structure is made up of β strand and coil regions (7~12,
23~26, 37~44, 51~56, 64~68, 78~83, 91~94 and
107~111 positions are mainly β strand structure). The
rest of the portion is helical structure interspersed with
coil regions. The structure is presented in Figure 1.
Figure 1 Model structure of SoxY protein from
A.Vinosum
Note: With distinct secondary structure showing as α-helix, β
sheet and random coil
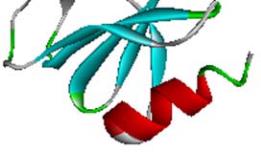
1.2 Description of the structure of SoxZ
The modelled structure of SoxZ is a 104 amino acid
residue long protein. The predicted structure is very
similar to the structure of the SoxZ protein from the
SoxYZ complex from
Paracoccus denitrificans
(PDB
code: 2OXG; Z chain for SoxZ). The protein is mainly
composed of β strand and coil structure. The structure
is presented in Figure 2.
Figure 2 Model structure of SoxZ protein from
A.Vinosum
Note: With distinct secondary structure showing as α-helix, β
sheet and random coil
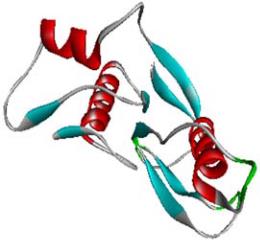
1.3 Description of the structure of C-terminal
domain of SoxB
The modelled protein structure of 5'-nucleotidase,
C-terminal domain of SoxB is a 138 amino acid
residue long protein. The predicted structure is very
similar to the structure of the SoxB.
Protein of Termus Thermophilus Sulfate Thiohydrolase
(PDB code: 2WDC; A chain for Sox B domain).The
residues show conformational adaptability towards helix,
β strand and coil conformations. There are four helical
and five βstrand regions in the modeled protein
structure (14~24, 63~68, 71~81,114~122 as helical
structure and 26~31, 36~40, 46~50, 95~99 and
133~136 as β strand structure). The helical and β
strand regions mainly interspersed with coil regions.
The structure is presented in Figure 3.
Figure 3 Model structure 5’-nucleotidase, C-terminal domain of
SoxB protein from
A.Vinosum
Note: With distinct secondary structure showing as α-helix, β
sheet and random coil
Computational
Molecular Biology

