基本HTML版本

Computational Molecular Biology 2015, Vol. 5, No. 1, 1-13
http://cmb.biopublisher.ca
5
So it maybe relates to hemoglobin sequences are well
conserved in evolution and between species. In some
parts of ß-chain sequences in the
Camelus
family
(
Camelus bactrianus
,
Camelus dromedarius
and
Camelus ferus
) the AA of H, D, N, H, G, S, K, V and
D were substituted by Q, E, A, L, A, D, N, E and E in
Equus caballus
respectively or the AA of S, G, D, N,
H, G, S, K and R in
Camelus
family were substituted
by T, P, E, S, T, A, G, N and Q in
Homo sapiens
respectively. So, differentiation of amino acid in the
species occurred during mutation into same
polypeptide which is major factor concerning to
hemoglobin efficiency in creatures. So, mutations
have played an important or unique role in causing
animals adapting to different environmental situations.
Lin et al., 1976 are declared that between adult camel
hemoglobin and adult human hemoglobin six amino
acid differences were found in the N-terminal 20
amino acid residues of the α-chain, at residues: 4, 5,
12, 14, 17, and 19; eight amino acid substitutions were
found in the ßchain at positions: 4, 5, 6, 9, 12, 13, 16,
and 19. Substitutions at α5 Ala → Lys, and ß19 Asn
→ Lys, increase the net positive charge of camel
hemoglobin by two, while other substitutions result in
no meaningful differences (Lin et al., 1976).
Pieragostini et al., 2010 presented an adaptive
significance of the hemoglobin variants with
hematological
patterns
in
ruminant
breeds
(Pieragostini et al., 2010). Braunitzer et al., 1980
declared five and two AAs (amino acids) in HBA and
HBB of Camelus ferus are exchange versus lama
respectively. As well as they deduced the AAs
sequence of Camelus ferus and dromedarius is
identical (Braunitzer et al., 1980). However, we
hypothesized that substitution of AAs into polypeptide
structure by protein engineering or cloning is an
appropriate approach to how increasing resistivity of
hemoglobin to glycosylation and subsequently to
diabetes mellitus.
2.2 Phylogenetic analysis
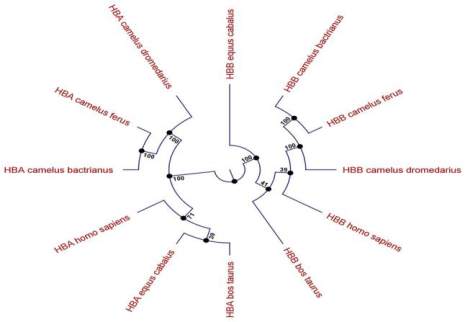
As shown in circular cladogram Figure 2 HbA in
Camelus bactrianus
and
Camelus ferus
were in the
same sub cluster with 100% bootstrapping near alpha
subunit of Camelus
dromedarius
. It also shown for
HBB in
Camelus
family (Figure 2). As phylogenetic
distance of
Camelus bactrianus
and
dromedarius
Figure 2 Circular cladogram by UPGMA. The evolutionary
history was inferred by using the UPGMA method based on the
JTT matrix-based model. The bootstrap consensus tree inferred
from 1000 replicates is taken to represent the evolutionary
history of the taxa analyzed. Initial tree(s) for the heuristic
search were obtained automatically as follows. The analysis
involved 12 amino acid sequences
α-chains versus
Homo sapiens
were more than
Bos
taurus
and
Equus caballus
(Figure 2). Phylogenetic
tree revealed ß-chain (HBB) of
Camelus bactrianus
,
Camelus dromedarius
and
Camelus ferus
were in the
same sub cluster with 95% bootstrapping and were
55% bootstrapping close to HBB of
Equus caballus
.
The based on circular cladogram Figure 2 HBA in
E
quus cabalus and
Bos tauurus
were in the same sub
cluster with 39% bootstrapping but far from HBA
Homo sapiens
slightly. Phylogenetic analysis had
demonstrated, every hemoglobin subunits are
conserved in species and it depends on particular
function and performance of protein.
2.3 Whole protein composition
Whole protein composition of HBA and HBB were
the same in the Camelus family but and was different
from other species entirely (Table. 2). The based on
Table 2, isoelectric point (p
I
) of hemoglobin in
Camelus
appeared to be more than either that of
Homo
sapiens
,
Bos tauurus
and
Ecuus cabalus
(Table 2). In
fact p
I
for hemoglobin subunits in camel is basic
rather than human. Also hemoglobin subunits in camel
contain more positively and negatively charged amino
acids than either Homo sapiens. The basic effects of p
I
in camels HBB is more than HBA (Table 2), thus we
imagine it’s related to high mobility of camel HBB

