Cancer Genetics and Epigenetics 2018, Vol.6, No.5, 33-39
34
1 From Cytosine to Reads: Second Generation Sequencing Based on Bisulfite Conversion
Conventional sequencing methods cannot distinguish between 5mC and unmethylated cytosines. Therefore, the
first step in detecting methylated cytosine in a DNA sequence is to distinguish 5mC from cytosine. Since the
discovery that sodium bisulfite can convert ordinary cytosine into uracil (the intermediate product is
5,6-dihydrouracil-6-sulfonate), the DNA methylation detection technology based on bisulfite conversion is
considered the gold standard for methylation detection, because it can accurately quantify the methylation status
of a single cytosine.
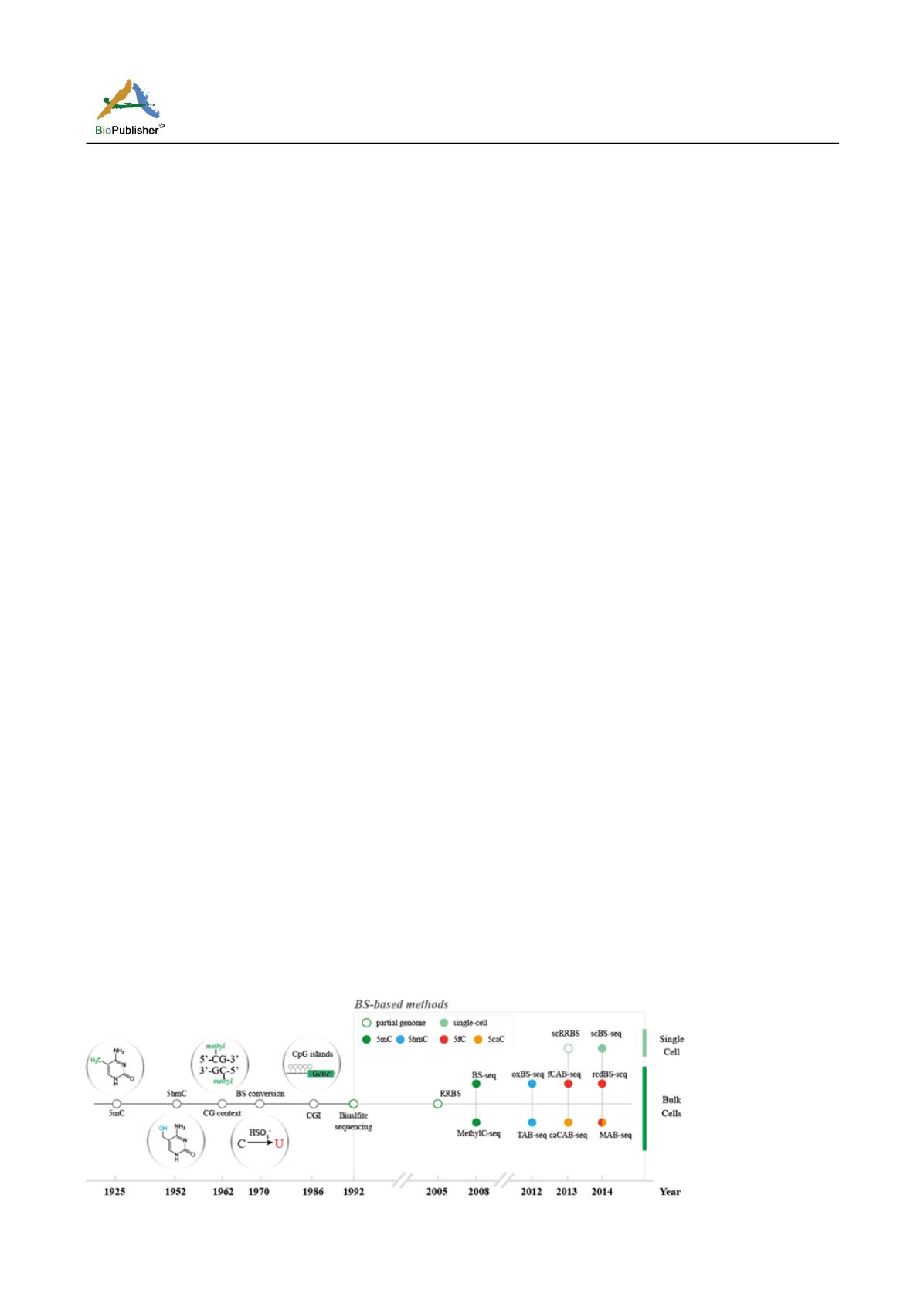
The bisulfite sequencing developed by Formmer et al. in 1992 can accurately recognize methylated cytosine on
the target sequence (Noble, 2009), but it usually only detects methylation status at dozens of CpG sites. The
development of next-generation sequencing has made it possible to detect the methylation status of whole-genome
cytosine. In March 2008, Shawn J. Cokus et al. examined the genome-wide DNA methylation profile of
Arabidopsis thaliana
, and its detection technique was named BS-seq. In May of the same year, Ryan Lister et al.
also published a whole genome DNA methylation sequencing of
Arabidopsis thaliana
, and its detection
technology was named MethylC-seq (Dudley and Butte, 2009). In addition to the slightly different methods of
building library between these two technologies, the core of these technologies is bisulfite conversion combined
with next-generation sequencing. Because it detects genome-wide cytosine methylation, it is collectively referred
to as Whole Genome Bisulfite Sequencing (WGBS), which is distinguished from RRBS (Reduced Representation)
which tends to detect CpG-rich regions (Bisulfite Sequencing) (Hackett et al., 2013).
There are four main steps in the detection of methylated cytosine by the WGBS technique: (i) Using ultrasound to
break DNA into short fragments. (ii) Treatment of the fragment with bisulfite: Bisulfite converts C to C while
5mC remains unchanged. (iii) PCR amplification of the fragment. (iv) The sequence of the base of the fragment
was detected using a sequencer. The methylation status of cytosine is inferred by comparison of the reads
produced by the sequencer with the reference genome.
In mammals, 5mC is the most, but not the only, cytosine modification. Recent studies have found that
5-methylcytosine (5mC) can be oxidized by Tet protein to 5-hydroxymethylcytosine (5hmC). While
5-hydroxymethylcytosine can be further oxidized by Tet protein into 5-formylcytosine (5fC) and
5-carboxylcytosine (5caC). These three cytosine modifications have been shown to be involved in the active DNA
demethylation process, and the TDG protein (thymine DNA glycosylase)-mediated base-excision repair process
can Reconvert 5fC/5caC to normal cytosine.
Conventional WGBS technology cannot detect three other cytosine modifications: bisulfite conversion does not
work for 5hmC and 5mC, while both 5fC and 5caC are converted to uracil. Considering these, the researchers
designed a series of variants of bisulfite sequencing technology to detect these three cytosine modifications
associated with DNA demethylation. For example, oxBS-seq (Lu et al., 2015) and TAB-seq are capable of
detecting whole-genome 5-hydroxymethylcytosine. fCAB-seq (Blaschke et al., 2013) and redBS-seq (Hu et al.,
2014) are capable of detecting 5-formylcytosine, caCAB-seq (Kohli and Zhang, 2013) can detect
5-carboxycytosine. In addition, MAB-seq (Klengel et al., 2013) can detect whole-genome 5fC/5caC (Figure 1).
Figure 1 Sequencing method based on bisulfite conversion


