Basic HTML Version




Molecular Plant Breeding 2011, Vol.2, No.15, 101
-
108
http://mpb.sophiapublisher.com
104
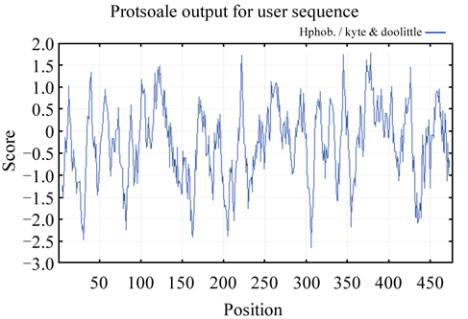
Figure 3 Hydrophobicity and hydrophilicity analysis of rice
rbcL
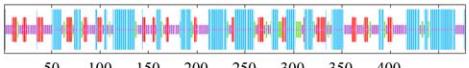
1.5 The rbcL secondary structure analysis of plants
The rbcL polypeptide secondary structure of
Oryza
sativa subsp. japonica
was detected with SOPMA
(Geourjon and Deléage, 1995). Alpha-helix and
random coil are the principal structural elements in
rbcL polypeptide of
Oryza sativa subsp. japonica
, and
extended strand and β-turn occupy a little scale, which
intersperse among the whole protein (Figure 4).
According to the statistic assay consequence, the
proportions of α-helix, extended strand, β-turn and
random coil in the rbcL secondary structural
components of
Oryza sativa subsp. japonica
are
40.25%, 16.56%, 9.85% and 33.33%, respectively.
Likewise, similary distributive regularity of the four
kinds of secondary structural units in rbcL has been
detected in many other higher plants, such as
Zea
mays, Triticum aestivum, Nicotiana tabacum,
Medicago truncatula, Oncidium Gower Ramsey,
Pisum sativum, Citrus sinensis
.
Figure 4 Secondary structure analysis of rice rbcL
1.6 Comparisons among the rbcL nucleotide
sequences and deduced amino acid sequences from
different plants
The homological comparisons of rbcL nucleotide
sequences and deduced AA sequences were
accomplished by online tool BLAST in NCBI
(Alcschul et al., 1997). The results indicate high
homologies of rbcL nucleotide sequences between
Oryza sativa subsp. japonica
and other higher plants
which are comprised of
Triticum aestivum, Nicotiana
tabacum, Lolium perenne, Medicago truncatula,
Oncidium Gower Ramsey, Calycanthus floridus var.
glaucus, Podocarpus macrophyllus
. The identities are
96%, 88%, 96%, 87%, 91%, 88%, 85%, respectively.
The identities of further deduced AA sequence
comparisons using BLASTp are 97%, 93%, 97%,
93%, 96%, 94%, 94%, respectively, while the
similarities of that are separately 99%, 97%, 99%,
97%, 98%, 97% and 98%. Relative to the results of
nucleotide sequence comparisons by BLASTn, higher
similarities can be discovered in the AA sequence
comparisons between
Oryza sativa subsp. japonica
and other six plants. Also, analogous results were
obtained in the comparisons of rbcL nucleotide
sequences and deduced AA sequences between
Oryza
sativa subsp. japonica
and a great number of other
higher plants, such as
Zea mays, Arabidopsis thaliana,
Pisum sativum, Citrus sinensis, Phalaenopsis
aphrodite subsp. formosana
. The regularity, which
exhibited higher and broader similarities in rbcL AA
sequences than in nucleotide sequences among various
plants, is supported by above-mentioned results.
The multiple alignment analysis of rbcL AA sequences
among
Cathaya argyrophylla
,
Calycanthus floridus
var. glaucus
,
Nicotiana tabacum
,
Solanum
lycopersicum
,
Citrus sinensis
,
Arabidopsis thaliana
,
Allium cepa
,
Phalaenopsis aphrodite subsp.
formosana
,
Oryza sativa subsp. japonica
and
Zea
mays
was carried out using Clustalx (Higgins and
Sharp, 1988; 1989; Thompson et al., 1997;
Jeanmougin et al., 1998) and DNAMAN software.
Super identities of rbcL AA sequences were illustrated
among higher plants, regardless of between
gymnosperm and angiosperm, or between dicotyledon
and monocotyledon (Figure 5). It was demonstrated
that there are exceeding conservatism and homologies
among higher plant rbcLs.
1.7 The molecular systemic evolution analysis on
rbcL DNA sequences from plants
The homologous evolutionary relationship among

