Basic HTML Version
Molecular Pathogens
4
kingae
genome that plays an important role in
initiation of inflammation and thereby increases the
chances of invasion (Kehl-Fie et al., 2007).
Kingella
kingae
has been reported to produce a polysaccharide
capsule, early during an infection mostly among
strains colonizing in the respiratory mucosa of very
young children (< 3 years) which indicates that the
immune system plays an important role in the
colonization and invasion of
Kingella kingae
, where
ineffective immune responses of children are not
sufficient to resist colonization and later invasion
(Porsch et al., 2012, Yagupsky et al., 2011)
immunological responses to invasive
Kingella kingae
infections has not been completely understood, but
studies from the past have reported the presence of
circulating IgG antibodies acquired from mother
would help in resisting colonization and infection in
children aged below 6 months and that as age
increases till 24 months the susceptibility to invasive
infection also rises (Slonim et al., 2003).
Microbiological, Laboratory Identification,
Confirmation and Antimicrobial Susceptibility
Testing
Kingella kingae
are facultatively anaerobic gram
negative bacilli, which on primary isolation appear as
cocci (resembling
Neisseria
spp.) and coccobacilli
(resembling
Moraxella
spp.) later on showing
bacillary forms (Ramana and Mohanty, 2009) (Figure
1). Though not strictly fastidious,
Kingela kingae
takes up to 48 hours for growth from clinical
specimens and on trypticase soy agar with added
blood (sheep blood agar), produces 1~3 mm pin point
to small β- haemolytic colonies, which are observed
sometimes to pit, spread or corrode the medium
(Kehl-Fie et al., 2009) (Figure 2).
Kingella kingae
are
catalase negative (differing with
Moraxella
which are
catalase positive), oxidase positive and non motile.
K.
kingae
are indole, urease negative and ferment glucose
and maltose only with production of acid and no gas.
K. kingae
can be differentiated from
Neisseria
spp by
using penicillin-G disc test, where
Kingella kingae
form elongated bacillary forms in the presence of
penicillin G disc (Yagupsky, 2004). Improved
isolation is achieved in case of strong clinical
suspicion, the clinical samples are incubated for at
least 48 hours and incubation in 5%~10% CO
2
chamber can improve the growth (Yagupsky, 2004).
On isolation, the regular biochemical reactions will be
sufficient to identify
Kingella kingae
. Primary
isolation from specimens can be improved by using
automated blood culture system (BACTEC
(BD-Becton Dickinson, Cockeysvillie, MD), Bac T
Alert systems (Yagupsky, 2004). Further confirmation
will be facilitated by API 20 system and Microscan
(Dade Behring, Germany) automated identification
and antimicrobial sensitivity systems depending on
their availability. Conventional PCR and real-time
PCR (RT-PCR) are the molecular methods that target
specific areas of DNA (cpn 60 and
RTX
genes) can be
used for confirmation and reducing the time for
diagnosis (Baticle et al., 2008; Ilharreborde et al.,
2009). Other methods including Multi-locus sequence
typing (MALT), SYBR green and TaqMan assays
have been used to sequence
rtxA
gene for
identification of
Kingell kingae
from various clinical
specimens using controls (
Kingella kingae
ATCC
23330) (Basmaci et al., 2012; Philippe et al., 2011).
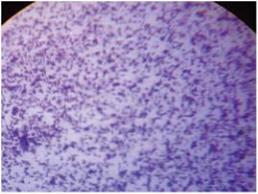
Figure 1 Gram’s stained smear of
Kingella kingae
showing
short gram negative bacilli
Figure 2 Colony morphology on Sheep blood agar after 48
hours of incubation showing 1~3 mm, pin-point to small
transcleucent colonies
Kingella kingae
, generally are susceptible to most of
the antimicrobial agents but there are reports of
production of beta lactamases (Yagupsky, 2004).
Molecular Pathogens

