基本HTML版本

Computational Molecular Biology 2014, Vol. 4, No. 15, 1-5
http://cmb.biopublisher.ca
4
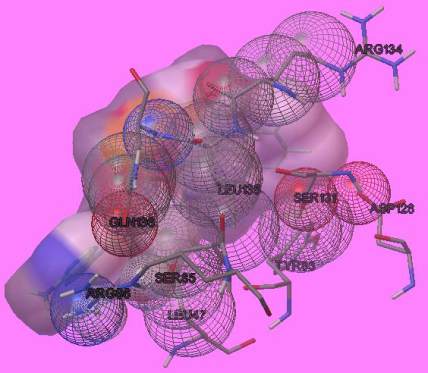
Figure 6 residues present at the active site of whole enzyme
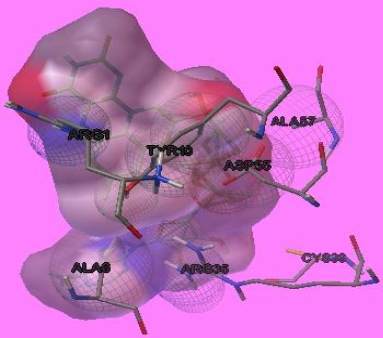
Figure 7 residues present at the active site of domain of the enzyme
parameters. The autodock and autogrid files were
generally run on python script after which the
interactions found by analyzing the dock.dlg files.
Less the binding energy stronger is the interaction.
However the binding energy is dependent on the
length of amino acids. The interaction result is shown
in Figure 8 and Figure 9 respectively.
Figure 8 FAD binding to whole enzyme
Figure 9 FAD binding to domain
2 Results
2.1 Retrieved sequence and finding the domain
region
The amino acid sequence of enzyme was retrieved
from uniprot with the uniprotID of Q4ADV8. The
protein contains 565 numbers of amino acids. The
domain region is found from 74-225. The information
about the domain region was found from both uniprot
and from protparam output.

