基本HTML版本


Computational Molecular Biology 2014, Vol. 4, No. 15, 1-5
http://cmb.biopublisher.ca
2
by using protparam tool. However the domain region can
be found from CDD (conserved domain database) too.
1.3 Alignment between whole protein sequence and
domain
Multiple sequence alignment is generally done to find
out the conserved residues among multiple sequences
(Thompson et al., 1994). From the alignment we
found the regions of common residues among the
domain part and whole sequence depending upon
which we could analyse the stronger binding region
where ligand (FAD) is binding. The alignment result is
shown in Figure 1.
Figure 1 multiple sequence alignment between domain and whole enzyme
1.4 Predicting the structures of domain and whole
protein
As the NMR crystallographic structure of enzyme is not
available at PDB, so the tertiary structure was generated
on the basis of homology modeling concept (Bilal et al.,
2013, Sungh et al., 2004, Sali et al., 1993).The models
for whole protein and domain parts were generated using
modeler 9.12 tool. The align2d.py, model-single.py,
evaluate-model.py files were run on the python script by
setting the template, target and the number of models to
be generated. In this study 10 models were generated for
both domain part and for whole enzyme sequence. The
best model was selected on the basis of lowest DOPE
score. The final generated models were visualized by
using visualizing tools i.e., pymol, discovery studio and
swiss-pdb viewer. Multiple visualiser tools were used to
understand the structures more properly (Table 1, Table 2).
The final models were shown in Figure 2 and Figure 3.
Table 1 Information found after visualizing the structures using
Pymol tool
Analysis
Protein structure Domain structure
Atom count
4233
1311
Formal charge sum
-6.0
0.0
Molecular surface area 52371.758A
0
16504.783 A
0
Solvent
accessible
surface area
23178.145 A
0
10648.261 A
0
Table 2 information found after visualizing the structures using
Yasara tool
Analysis
Protein structure
Domain structure
VDW radius
85.337
34.261
Beta factor
109.7
111.3
Mass of object
55813.069g/mol
17316.863g/mol
Stability of the object 971.13kcal/mol
286.78kcal/mol
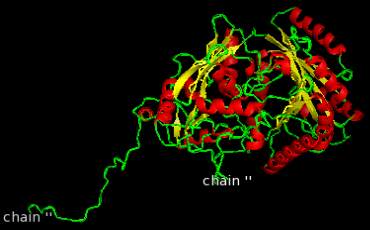
Figure 2 Model generated for whole enzyme
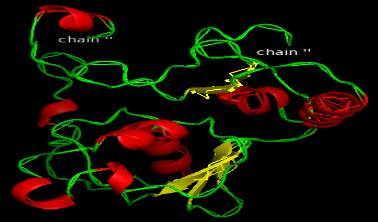
Figure 3 Model generated for domain

