基本HTML版本

Computational Molecular Biology 2014, Vol. 4, No. 13, 1-6
http://cmb.biopublisher.ca
3
1.3 Prediction of templates
The similarity search is generally done by using
BLAST tool and the protein-protein similarity search
is carried out by using the blastP tool. So the retrived
amino acid sequence was subjected to blastP against
PDB. From the result the suitable templates were
found for further study. The selected templates for
model building are given in Table 4.
Table 4
List of four templates used for homology modelling
Templates
Chain
Identity
Query cover E-value Organism
Molecule
1IZL
D
89%
100%
0.0
Thermosynechocuccus vulcanus PhotosystemII subunit psb A
4IL6
D
90%
96%
0.0
Thermosynechocuccus vulcanus photosystemQ (B) protein
3A0B
D
95%
91%
0.0
Thermosynechocuccus vulcanus photosystemQ (B) protein
3WU2
D
96%
90%
0.0
Thermosynechocuccus vulcanus photosystemQ (B) protein
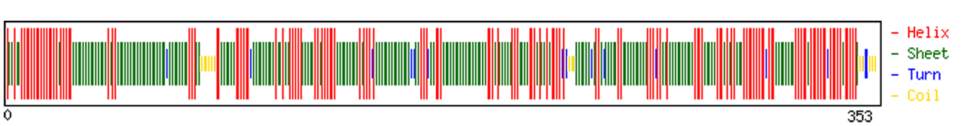
1.4 Secondary structure prediction of protein
The secondary structure of protein was predicted by
using CFFSP server from where the percentage of
helix, sheets and turns were found (Figure 1). The
detail information is given in Table 5.
Table 5 composition ofhelices, sheets, turns
Templates
Helices
Sheets
Turns
1IZL
36.8%
0.0
5.7%
4IL6
51.3%
0.0
3.4%
3A0B
55.2%
0.0
1.1%
3WU2
54.1%
0.0
6.2%
Figure 1Secondary structure ofPHOTOSYSTEMII D2 protein
1.5 Homology modelling
The 3D structures of the photosystemII D2 protein was
generated by using homology modelling concept, in
which four different templates were selected for model
building.The models were generated by using modeller
9.12 tool(Bilal et al., 2013, Singh et al., 2009). The
align2d.py, model-single.py and evaluate-model.py
files were rum on the python script by setting the target,
template and number of models to be generated. Here 5
models for each template were generated and the best
model was selected on the basis of lowest DOPE score.
The properties which were found after structure
visualisation is given in Table 6 and Table 7. the tertiary
atructures of protein is given in Figure 2-Figure 5.
Table 6 Result obtained from yasara tool
Templates
Beta factor
Stability of object
Minimized energy
VDW radius
1IZL
140.0
930.7kcal/mol
-0.737849
60.267A
0
4IL6
110.9
723.92 kcal/mol
-2.8532
53.888 A
0
3A0B
105.4
607.54 kcal/mol
-2.21035
53.484 A
0
3WU2
99.8
591.03 kcal/mol
-1.38159
52.549 A
0
Table 7 Result obtained frompymol tool
Templates
Atom count
Formal Charge sum
Molecular surface area
Solvent accessible surface area
1IZL
2804
-9.0
34263.043 A
0
21350.953 A
0
4IL6
2804
-9.0
34034.191 A
0
21938.918 A
0
3A0B
2804
-9.0
34332.879 A
0
21257.391 A
0
3WU2
2804
-9.0
34461.117 A
0
21316.729 A
0

