基本HTML版本

Computational Molecular Biology 2014, Vol. 4, No. 9, 1-6
http://cmb.biopublisher.ca
4
Table 5 Enzyme Code (EC) Classification
Species
Oxidoreducta---ses Transferases Hydrola-ses Lyases
Isomera--ses Ligases
Total
Arachis hypogaea
L.
301
614
431
78
51
71
1546
Cicer arietinum
L.
51
92
76
20
5
4
248
Phaseolus vulgaris
L.
110
232
147
23
20
31
563
Trigonella foenum-graecum
L.
148
149
179
34
38
27
575
Vicia sativa
L.
429
927
718
80
82
100
2336
2.4.3 Gene Ontology (GO) Classification
To functionally categorize various legume transcript
contigs, Gene Ontology (GO) terms were assigned to
each assembled transcript contigs. Transcript contigs
were grouped into GO functional categories
(http://www.geneontology.org), which are distributed
under the three main categories of Molecular Function,
Biological Process and Cellular Components (Table 6).
Table 6 Gene Ontology (GO) Classification
Species
Molecular Function
Biological Process
Cellular Components
Total
Arachis hypogaea
L.
4512 (43%)
3352 (32%)
2607 (25%)
10471
Cicer arietinum
L.
916 (40%)
734 (32%)
654 (28%)
2304
Phaseolus vulgaris
L.
1727 (47%)
1168 (31%)
829 (22%)
3724
Trigonella foenum-graecum
L.
2792 (28%)
4407 (43%)
2980 (29%)
10179
Vicia sativa
L.
7026 (37%)
5815 (31%)
5920 (32%)
18761
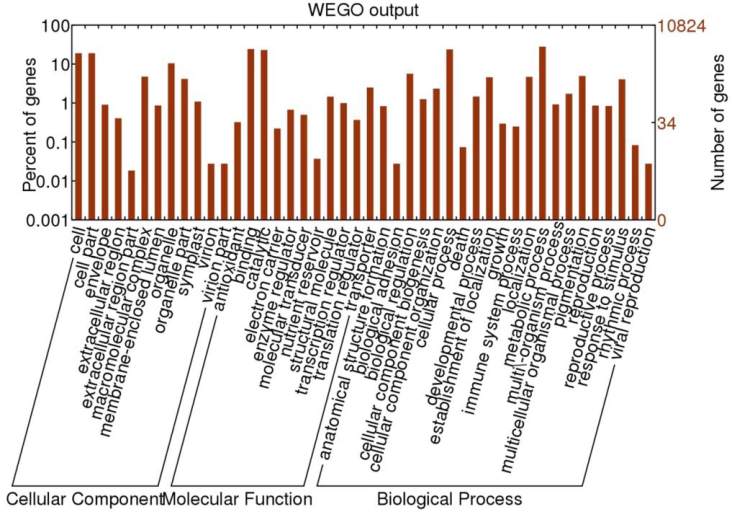
Figure 1 which is output of WEGO tool; it shows that,
Within the Molecular Function category, genes encoding
binding proteins and proteins related to catalytic activity
were the most enriched. Proteins related to metabolic
processes and cellular processes were enriched in the
Biological Process category. With regard to the Cellular
Components category, the cell and cell part were the
most highly represented categories. We found same in all
other legume species so we have considered only this
one figure for illustration of WEGO tool.
Figure 1 WEGO Tool Result of Arachis hypogaea L.

