基本HTML版本

Computational Molecular Biology 2014, Vol. 4, No. 7, 1-17
http://cmb.biopublisher.ca
6
number represents a lower bound of a species
secretome. Including other proteins predicted as likely
secreted and weakly likely secreted proteins, the size
of secretome certainly will be significantly increased,
but there would be an increase in the number of false
positives, i.e., non-secreted proteins in the set.
2.3
Relationship of lifestyle and secretome size in
different fungi
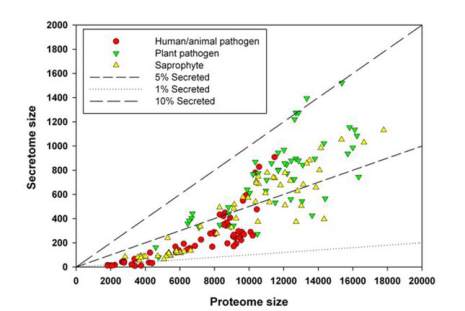
Similar to our previous analysis in FunSecKB work
(Lum and Min, 2011), the secretome size (Y) was
highly correlated with its proteome size (X) in a
species (r = 0.87) with a regression as Y = 0.081X -
271. (Figure 1). However, species having different
lifestyles showed differences in secretome size and
proportion of secreted proteins. Lowe and Howlett
(2012) examined the relationship between lifestyle
and secretome size and found that fungi with biphasic
lifestyle have a large proportion of secreted proteins
and animal pathogens have fewer genes than
saprophytes or plant interacting fungi do, and a lower
proportion of predicted secreted. In the work of Lowe
and Howlett (2012), the secretome prediction was
only used SignalP, and thus, its size may be over
estimated. Using the data we collected in this work,
we examined the relationship between fungal
lifestyles and their secretome sizes (Figure 1,
. As the data for each species
in the database contain redundant or duplicated
protein entries, we only used the proteins in datasets
of reference or complete proteomes compiled by UniProt
(http://www.uniprot.org/taxonomy/complete-proteome
s). We collected species having a complete proteome
and a lifestyle in the category of animal or/and human
pathogen, plant pathogen, and saprophyte. Some of
them may be classified into more than one category
and these entries are annotated (see
. In general agreement with Lowe and
Howlett (2012) reported, human and animal pathogens,
including entomopathogens and some nematode
killing fungal parasites have a relatively smaller
proteome size – the majority of them have <12000
protein sequences, some of them are known as
Microsporidian parasites having a genome encoding a
total of 2000 - 4000 proteins, with less than 1% of
them being secreted (Figure 1). The proportion of
secreted proteins varied from 0.3 to 7.9% with an
average of 2.8% in human/animal pathogens. On other
hand, plant pathogens and saprophytes have much
more variable proteome sizes from ~ 4000 to 18000
and a relatively higher proportion of secreted proteins,
though variable, from 1.3 to 7.1% with an average of
4.2% in saprophytes and from 1.7 to 10.5% with an
average of 6.3% in plant pathogens. Clearly, these
results show that secretome size is one of the
important determining factors in controlling fungal
lifestyles. However, as species having a similar size of
secretome may have different lifestyles, the
composition within each secretome may play a more
critical role in determining its lifestyle in each species.
Figure 1 Relationship between proteome size and secretome
size in fungal species having different lifestyles
2.4 Functional analysis of fungal secreted proteins
To provide an overview of the functionalities of all
fungal secreted proteins, we carried out Gene
Ontology (GO) analysis. The secreted protein set
including curated and predicted highly likely secreted
proteins only was used to search the
UniProtKB/Swiss-Prot dataset with BLASTP with a
cutoff E-value of 1e-10. GO information was retrieved
from UniProt ID mapping data (http://www.uniprot.
org/downloads) and analyzed using GO SlimViewer
with generic GO terms (McCarthy et al., 2006). GO
biological
process
and
molecular
function
classification of the secretomes are summarized in
Table 3. Molecular function classification revealed
that fungal secreted proteins consist of a large number
of hydrolases (~33.7%), proteins having ion binding

