Basic HTML Version


Molecular Plant Breeding Provisional Publishing
Molecular Plant Breeding 2012, Vol.3, No.
6
, 57
-
62
http://mpb.sophiapublisher.com
58
sensitive to factors such as the day-night circle, which
limits generation of different kinds of starches
(Jobling, 2002).
Expression of
E. coli glg
B in amylose-free potato led
to generation of tuber starch with low molecular
weight and increased branch points (Anne et al., 1996).
Amylose is pe-required for formation of alpha 1-6
branch chain in starch; however, much less amyloses
were used as substrates in amylose free potato. Branch
enzymes, encoded by native potato SBE and
E. coli
glg
B expressed in potato, work with much less
amyloses as substrates. This resulted in an unstable
reaction catalyzed by the branch enzymes in tubers
since both branch enzymes were sensitive to the day
and night cycle.
With a relative constant (not much less in amylose
potato) supply of amyloses as substrates, shortage of
substrates for alpha 1~6 branch as it is in amylose free
potato could be avoided. In other words, branch
enzyme activity was increased by expression of
glg
B
in wild type potatoes rather than in mutants. Starch
molecular weight and other property could be
improved, but it is still unknown what aspect of starch
was improved when branch enzyme was enhanced in a
wild type potato.
In this study,
glg
B, driven by 35S promoter is
expressed in the two wild types of potatoes. It was
found that
glg
B expression showed a genotype
specific effect on starch viscosity. Results showed
that the starch tuber starch viscosity in
glg
B
transformed lines was 15 time higher than that in the
control. Measurement and analysis of the starch
indicate that
glg
B can be expressed in different
potato genotypes, which result in different starch
types with improved starch viscosities as high as 15
times as the control group.
1 Results
1.1
glg
B isolation
With
glg
B specific primer, a 2 184 bp product was
amplified as shownin Figure 1A. The product was
ligated into pGEM-T and sequenced. pG-gB+ and
BL-gB- were recognized after sequence alignment.
With the
glg
B sequence BLAST was done in the
Nucleotide database in NCBI. It was found that
glg
B
sequence isolated from this study is 99% similar to the
glg
B sequence in the JM109 genome recorded in the
database (Welch et al., 2002). Putative polypeptide of
the
glg
B is made up of 728
-
amino acid. After aligning
it with other
glg
B sequences from other bacterial
strains, it was found that there are 3 specific amino
acid mutation sites in the putative polypeptide, which
are K39E, V71A and I248V. The sequence was
registered in NCBI nucleic acid database with
accession number EU44744.
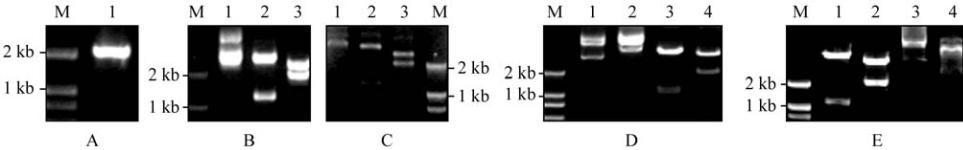
Figure 1
glg
B isolation and constructing recombinant plasmids
Note: A:
glg
B DNA from PCR; M: DNA molecular weight marker; 1: 2 186 bp
glg
B DNA; B: Digestion of recombined plasmid
pG-gB+; M: DNA molecular weight marker; 1: Recombined plasmid pG-
g
B
+
DNA; 2: Products of recombined plasmid pG-gB+
digested by
Bam
H
Ⅰ
; 3: Products of recombined plasmid pG-gB+ digested by
Nco
Ⅰ
and
Sac
Ⅰ
;
C
: Digestion of recombined plasmid
pG-gB-; M: DNA molecular weight marker; 1: Recombined plasmid pG-gB- DNA; 2: Products of recombined plasmid
pG-gB-
digested by
Bam
H
Ⅰ
; 3: Products of recombined plasmid
pG-gB- digested by
Nco
Ⅰ
and
Sac
; D: Digestion of recombined plasmid
pE-gB+; M: DNA molecular weight marker; 1: Plasmid pET-28c DNA; 2: Recombined plasmid pE-gB+DNA; 3: Products of
recombined plasmid
pE-gB+ digested by
Bam
H
Ⅰ
; 4: Products of recombined plasmid
pE-gB+ digested by
Xba
Ⅰ
and
Pst
Ⅰ
; E:
Digestion of recombined plasmid pE-gB-; M: DNA molecular weight marker; 1: Products of recombined plasmid pE-gB- digested by
Bam
H
Ⅰ
; 2: Products of recombined plasmid
pE-gB- digested by
Xba
Ⅰ
and
Pst
Ⅰ
; 3: Recombined plasmid pE-gB-; 4: Plasmid
pET-28c

