Basic HTML Version


Molecular Plant Breeding Provisional Publishing
Molecular Plant Breeding 2012, Vol.3, No.5, 50
-
56
http://mpb.sophiapublisher.com
55
genotypes. Transgenic plants were selected on
selection regime having at 10 mg/L basta and 5 mg/L
basta for genotype S-2003-us-359 and S-2003-us-127
respectively.
3.4 Preparation of gold particle
Gold particles (40 mg) with an average size (0.6 µM)
were suspended in 1ml of 96% ethanol. Centrifugation
was done at 4 200 rpm for 1 minute. Supernatant was
removed followed by the addition of 1 mL (96%)
ethanol. Resuspended the particles again for short
time and repeated three times. Particles were
washed in 1 mL ultra pure water for three times.
Resuspended the particles again in 1 mL ultra pure
water after last centrifugation treatment. Aliquots of
50 µL were formed and stored aliquots at
-
80
℃
.
3.5 Bombardment of calli with
bar
gene and
selection of putative transformed plants
The plasmid DNA having
bar
gene (Figure 8) was
precipitated on to 0.6 μ gold particles. DNA coated
gold particles were bombarded (PDS 1000/He). The
bombarded calli of genotype S-2003-us-359 and
S-2003-us-127 were transferred to Regeneration
selection media having 5 and 10 mg/L basta respectively,
incubated for 8~16 hrs dark and light condition at
(26±1)
℃
. Only those plants were survived on basta
which have
bar
gene and others were died.
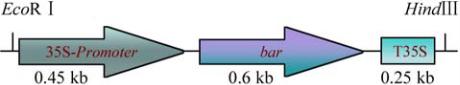
Figure 8 PCa
bar
Expression cassette with 35S promoter,
bar
and T35S terminator and restriction sites
3.6 DNA extraction
Leaf tissues (200 mg) were ground in liquid nitrogen
700 µL Extraction buffer was added in each reaction
tube followed by the addition of 800 µL phenol
chloroform Isoamylalcohol (25:24:1). Centrifugation
was done for 3 minute at 5 000 rpm at 4
℃
.
Supernatant was taken followed by the addition of
1/10 of sodium acetate into each tube. Equal volume
of Iso-propanol was added into each tube. Centrifugation
was performed at 13 200 rpm for 15 minutes.
Supernatant was removed. Washing of pellets with
80% ethanol was done, followed by air drying the
pellets. The pellet was dissolved in R40 (40 μg/mL
RNAse A in 1× TE, pH 8.2). DNA quantity and
quality was checked with nanophotometer and by
running on 0.8% agarose gel.
3.7 PCR analysis
Plants which were survived on basta were analyzed
with PCR (Polymerase chain reaction) analysis.
Putative plants which were survived on basta were
compared with wild type. PCR was carried out in a
25 µL reaction volume, containing 13.75 µL d
3
H
2
O,
2.5 µL
10
×
Taq
buffer, 2.5 µL MgCl
2
, 1 µL dNTPs,
0.25 µL
Taq
DNA Polymerase, 1 µL of each primer
(reverse and forward primers flanking the bar gene)
and 3µl Template DNA. The sequences of reverse and
forward primers of
bar
gene are as follows.
Table 3 Primers used for the amplification of
bar gene
for
transgenic confirmation
Primer name Primer sequence (5
'→
3')
Length
IQR/BAR-1 GAGACCAGTTGAGATTAGGCC
21
IQR/BAR-2 ATCTGGGTAACTGGCCTAACT
21
3.8 PCR (RAPD) amplification profile
PCR amplification was done by incubating the each
DNA samples at 95
℃
for 3 minutes, then 35 cycles
comprising of denaturation at 95
℃
for 1 minute,
annealing of primers at 58
℃
for 1 minute and
extension at 72
℃
for 1 minute. The final extension
was carried out at 72
℃
for 10 minutes. Resolving of
PCR product was done by gel electrophoresis using
ethidium bromide staining solution. Agarose gel
electrophoresis separates macromolecules on the basis
of charge, size, or other physical properties. PCR
product were resolved on 0.8 Agarose in 0.5
×
TAE.
After electrophoresis, gels were photographed using gel
documentation system, and gel pictures were saved.
Reference
Akama K., Puchata H., and Hohn B., 1995, Efficient
Agrobacterium
mediated transformation of Arabidopsis thaliana using the bar gene as
selectable marker, Plant Cell Rep., 14: 450-454 http://dx.doi.org/10.1007/
BF00234053
Anjum N., Ijaz S., Rana I.A., Khan T.M., Khan I.A., Khan M.N., Mustafa G.,
Joyia F.H., and Iqbal A., 2012, Establishment of an in vitro
regeneration system as a milestone for genetic transformation of
sugarcane (
Saccharum officinarum
L.) against
Ustilago scitaminea
,
Bioscience Methods, 3(2): 7-20
Cao M. X., Huang J.Q., He Y.L., Liu S.J., Wang C.L., Jiang W.Z., and Wei

