Basic HTML Version



Cotton Genomics and Genetics 2012, Vol.3, No.1, 1
-
7
http://cgg.sophiapublisher.com
4
appearing in the non-transgenic cotton. There were
135 individuals in 281 transgenic plants belonging to
transgenic Zhong35, the rest 128 individuals were
Transgenic Juanmian 1 hao. It might be proved that
the target gene had been integrated into the genome of
the cotton plant with a certain genetic stability.
The PCR product of the marker
Bar
gene was
amplified. The electrophoresis results showed that
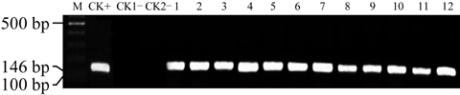
transgenic plants had a clear band with 146 bp in
length that located in the same position as the plasmid
amplification products. There were 153 individuals of
Zhong35 and 128 individuals of Juamin 1 Hao,
whereas no band amplified in non-transgenic plants
(Figure 3). It is obvious that the number of the
detected transgenic plants was consistent with the
PCR assay for
SNC1
gene.
1.6 T
1
generation of transgenic cotton plants
identified by RT-PCR
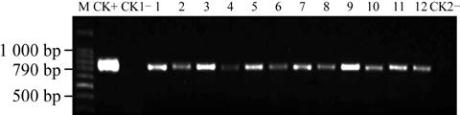
When the transgenic cottons grew up with the six-leaf
stage, total RNAs of 281 individuals of the T
1
generation with SNC1 gene were extracted from
leaves for RT-PCR detection. The target gene
SNC1
with the band of 790 bp in length at the transgenic
plants were detected with the same size as the positive
control, which indicated that the
SNC1
gene in the T
1
generation should express in the transgenic cotton
plant in the RNA level (Figure 4).
1.7 Identification of disease resistance of transgenic
cotton T
1
generation plants by
in vitro
infection
A certain concentration of cotton Fusarium wilt strain
(
Fusarium oxysporum
) was inoculated in the culture
pot to cause cotton seedling wilt occurring within a
week. The rates of disease incidence were recorded
every 5 days until the incidence of disease has been
Figure 2 Detection of
SNC1
gene in part of the transgenic
plants by PCR
Note: M: 100 bp DNA ladder; CK+:
SNC1
plasmid; CK1
-
:
Non-transgenic plants; 1~12: Transgenic plants; CK2
-
: Blank
(water)
Figure 3 Detection of selective marker
Bar
in part of the
transgenic plants by PCR
Note: M: 100 bp DNA ladder; CK+:
SNC1
plasmid; CK1
-
:
Non-transgenic cotton; 1~12: Transgenic cotton; CK2
-
: Blank
(water)
Figure 4 Detection of some
SNC1
transgenic plants by RT-PCR
Note: M: 100 bp DNA ladder; CK++: SNC1 plasmid; CK1
-
:
Non-transgenic cotton; 1~12: Transgenic cotton, CK2
-
: Blank
(water)
stable in the 21 days. The rate of incidence showed
that in the 21days, the incidence rate of Juanmian 1
hao as control had 66.7%, while the incidence rate of
transgenic Juanmian 1hao had 37.5% that is about half
of the control; Whereas the incidence rate of Zhong35
as control had 50.0%, while the incidence rate of
transgenic Zhong35 had 22.2% that is less than half of
the control. This result indicated that the introduction
of the exogenous gene SNC1 into cotton varieties
should acquire obvious disease-resistance compared
with non-transgenic cotton varieties, while genetically
modified Zhong3 had much more significant disease
resistance effect than that of the transgenic cotton
Juanmian 1 hao (Table 4; Figure 5).
2 Discussion
Although the Agrobacterium-mediated method is a
more mature approach of transgene, only a few
in
vitro
culture materials of varieties can be regenerated
because of long experimental period and limits of
genotypes in somatic explant regeneration
in vitro
.
Cotton shoot apex culture for regeneration is relatively
easy due to using complete meristem. Since Renfroe
et al reported that they obtained the regenerated plants
using cotton shoot meristem, many scholars at home
and abroad reported the evidences of transgenic plant

