Computational Molecular Biology 2015, Vol.5, No.6, 1-4
3
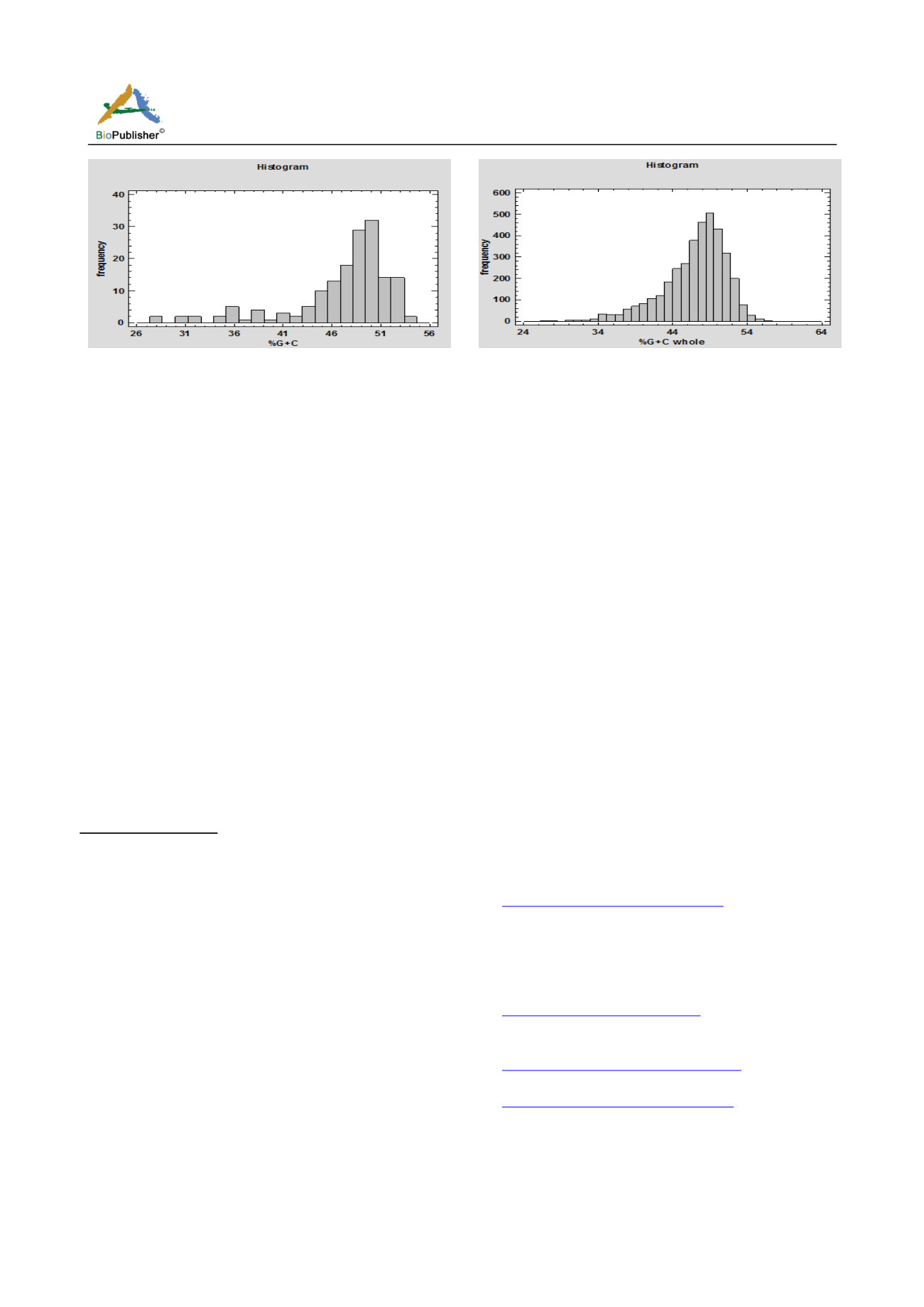
Figure 1 Distribution of GC content among the genes of VGS
We consider organism whether it is bias or unbiased in
relation with its codon usage if highly expressed genes
shows a different distribution of synonymous codons
from that in other genes in the genome. Several
methods were proposed to assess the extent of codon
usage bias at an organism scale, which determine the
organism as biased or unbiased. NC
diff
is a most
widely used measure to judge whether the organism is
biased or unbiased. Here we use the measure to
evaluate the extent of codon usage bias of
Vibrio
cholera N16961.
The difference of average NC values
of the ribosomal protein coding genes and the average
NC values of the rest of the genes in the genome are
referred to as NC
diff
. Organism having high value and
low value of NC
diff
exhibit large and small extents of
codon usage bias respectively.
NC
diff
was obtained by:
NC(all) – NC(rib)
NC(all)
The NC
diff
value of this bacteria is very low which
denotes that this bacteria shows a very small extant of
codon usage bias in its whole genome.
2.3 GC content of VGS and whole genome
Total GC content of pathogenic gene set and the
whole genome gene set (devoid of pathogenic gene)
were measured and it has been found that there is
uniform distribution GC between this two gene set,
which implies that pathogenic gene set not influenced
by mutational pressure and other factors and there is
hardly any chance of horizontal gene transfer into the
pathogenic gene set of vibrio cholera. Total GC
content of whole genome and VGS (pathogenic set)
Figure 2 Distribution of GC content among the genes of whole
genome
respectively 47% and 48%, the variation of GC
content between this two gene set is negligible.
3 Conclusion
From this study it can be observed that there is no any
difference in codon usage pattern of VSG and genome
of
Vibrio cholera N16961
as well as pathogenic gene
set share the same pattern of distribution of
nucleotide composition with the whole genome.
The selection force i.e. mutational pressure are
influencing in a similar fashion in both the gene
set of
Vibrio cholerae N16961
. This finding
also support that there is hardly any genome
wide codon usage difference as we find that
extent of codon usage bias is very low
throughout the genome, which indicate that
there may be very least chance of codon usage
difference among the different functional
categorical genes in this pathogenic bacteria.
References
Blake P.A. and Olsvik Ø. (eds.), 1994, In Vibrio cholerae and Cholera:
Molecular to Global Perspectives, ASM Press, pp. 293-295
Lawrence J.G. and Roth J.R.,1996, Selfish Operons: Horizontal Transfer
May Drive the Evolution of Gene Clusters, Genetics, 143(4):
1843–1860
Ochman H. and Moran N., 2001, Genes lost and genes found: evolution of
bacterial pathogenesis and symbiosis, Science, 292: 1096–9
Liu Q.P., 2006, Analysis of codon usage pattern in the radioresistant
bacterium Deinococcus radiodurans, BioSystems, 85(2): 99-106
Read A.F., 1994, The evolution of virulence, Trends Microbiol, 2: 73–81
Waldor M.K., Colwell R., and Mekalanos J.J., 1994, The Vibrio cholerae
O139 serogroup antigen includes an O-antigen capsule and
lipopolysaccharide virulence determinants. Proc. Natl Acad. Sci. USA
91, 91(24): 11388–11392


