Basic HTML Version





Bt Research
15
Worldwide, there are several reports of field evolved
resistance in
H. armigera
against
Cry
toxins (Van
Rensburg, 2007, Dhurua and Gujar, 2011). Resistance
has been reported to develop in Australia (Mahon et
al.,
2007); USA (http://www.epa.gov/ oppbppd1
/biopesticides/ pips/Bt_corn_refuge_2007.html.); Arizona
state of USA (Van Rensburg, 2007, Tabashnik et al.,
2008); China (Liu et al., 2008). Also, in India
resistance scenario is becoming a matter of great
concern, pink bollworm has already developed
resistance against Cry1Ac (Dhurua and Gujar, 2011,
Tabashnik and Carriere, 2010). Similarly,
H. armigera
from Bathinda and Muktsar (Punjab) have reported to
develop resistance against Cry1Ac (Kaur and Dilawari,
2011). Apart from using various field adopted insect
resistance management (IRM) strategies, molecular
IRM strategies have potent genes/protein to overcome
the resistant population. It is a suitable alternative to
sustain the success of
Bt
transgenic technology in India.
The discovery of novel genes from native
Bt
can
overcome the unavoidable resistance development in
insects. The present study has explored the native
isolates of
Bt
to screen the presence of
vip
genes and
its diversity. This study also promises to yield more
potent strain of
Bt
than standard
Bt
. By exploring the
possibilities of getting new and more potent
Bt
vegetative insecticidal protein genes, we can
overcome or prolong by increasing insect resistance
against
Bt
toxins.
1 Results
1.1 Molecular screening of local
Bt
isolates
Four isolates viz. PDKV-08, PDKV-27, PDKV-28 and
NCIM-5112, showed amplification for
vip1
/
vip2
gene
with expected size of 742 bp (Figure 1).
vip1
/
vip2
proteins have prime future prospects due to potent
insecticidal activity against coleopteran insect pest.
Warren in 1997 reported 11.9% a bundance of
vip1
/
vip2
gene amongst 463
Bt
isolates. Shi et al.
(2006) also confirmed that these two genes are located
on single operon and co-expressed.
Twenty eight local isolates, eleven different
Bt
strains
and reference strain HD1 were surveyed for the
presence of
vip3A
genes. Amongst 40 strains 5 strains
viz., PDKV-08, PDKV-21, NCIM-5110, NCIM-5132
and HD-1 showed the presence of
vip3A
gene by
using
vip3A
(partial) primer (Figure 2A).
Figure 1 Screening of
Bt
isolates for the presence of
vip1
/
vip2
binary toxin gene
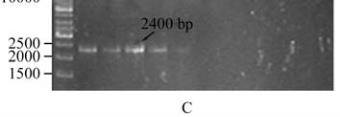
Figure 2 Screening of
Bt
isolates for the presence of
vip3A
genes
Note: A: 1~9 (1 kb DNA; PDKV-1; PDKV-3; PDKV-21;
NCIM-2513; NCIM-5110; NCIM-2514; NCIM-5123; HD-1);
B: 1~9 (1 kb DNA; PDKV-4; PDKV-8; PDKV-16; PDKV-20;
PDKV-21; NCIM-5132; NCIM-2130; HD-1); C: 1~12 (1 kb DNA;
PDKV-3; PDKV-8; NCIM-5110; NCIM-5132; HD-1; PDKV-6;
PDKV-14; PDKV-16; PDKV-20; PDKV-26; PDKV-28)
Bt Research

