Basic HTML Version



Genomics and Applied Biology
, 2013, Vol. 4 No.1 1-7
http://gab.sophiapublisher.com
abscisic acid inducible gene expression, and interact
and inactivate mitogen-activated protein kinase
(MAPK, Umbrasaite et al., 2010).
AtPP2C52 was clustered into Group E (Xue et al.,
2008). The interaction between the heterotrimeric G
proteins β subunit (AGB1) and AtPP2C52 has been
confirmed by Y2H analysis and an
in vitro
pull-down
assay in our previous work (Tsugama et al., 2012a).
Here we proved that
AtPP2C52
is expressed in almost
all the plant organs with a higher level in the vascular
and meristem. AtPP2C52 can interact with UMP1 and
RD21a as well as AGB1.
1 Results
1.1 Interaction between AtPP2C52 and AGB1
in
vitro
- 2 -
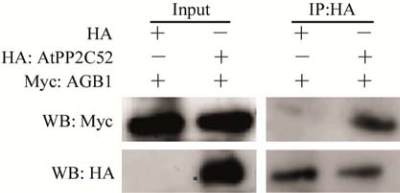
AtPP2C52 (GenBank Accession No.: NP_680572)
was identified as an AGB1-interacting protein
(Tsugama et al., 2012a). Coimmunoprecipitation
(Co-IP) was used to confirm the interaction of
AtPP2C52 and AGB1
in vitro
(Figure 1). Myc-tagged
AGB1
(
Myc:AGB1
)
, HA-tagged AtPP2C52
(
HA:
AtPP2C52) and HA epitope tag
(
HA) were syn-
thesized
in vitro
in a rabbit reticulocyte lysate system.
Either HA epitope tag or HA:AtPP2C52 was mixed
with Myc:AGB1, and then precipitated by anti-HA
antibody. Subsequently, G Sepharose was added. After
incubation, Myc:AGB1 in the elutant from the G
Sepharose was analyzed by immunoblotting using
anti-Myc antibody. Specific signals of Myc:AGB1
were detected only when AtPP2C52 was present
(Figure 1), indicating that AtPP2C52 interacts with
AGB1
in vitro
.
Figure 1 AtPP2C52 interacts with AGB1
in vitro
Note: HA epitope tag or HA-tagged AtPP2C52 (HA: AtPP2C52)
was immunoprecipitated with anti-HA antibody; Western-blot
using anti-Myc antibody revealed that anti-HA antibody
co-precipitated with Myc-tagged AGB1 (Myc:AGB1) in the
presence of HA:AtPP2C52 but not in the presence of HA
epitope tag
1.2 P
AtPP2C52
::GUS analysis
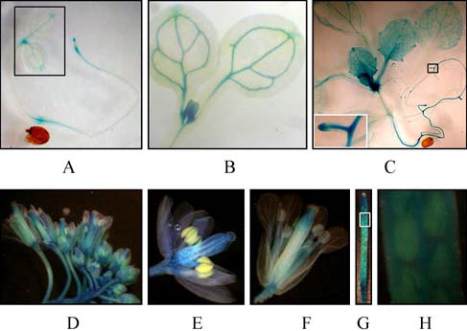
To analyze the temporal-spatial expression pattern of
AtPP2C52, transgenic plants expressing a promoter-
reporter fusion gene (P
AtPP2C52
::GUS) were used.
P
AtPP2C22
::GUS was expressed in almost all the plant
organs (Figure 2).
In 4-day-old seedlings, P
AtPP2C52
::GUS was predo-
minantly expressed in vascular, root tip and apical
meristem (Figure 2A; Figure 2B). In 3-week-old
plants, P
AtPP2C52
::GUS was evident in the whole plant
(Figure 2C). The expression of P
AtPP2C52
::GUS was
still higher in vascular and apical meristem in this
stage. In adult plants, P
AtPP2C52
::GUS was found in all
the organs of flower, excepting the anther (Figure 2E;
Figure 2F). The expression level of P
AtPP2C52
::GUS
was lower in the sporangia (Figure 2G; Figure 2H).
Figure 2 Temporal and spatial expression of
AtPP
2C52
Note: A: GUS staining showing the expression of
P
AtPP2C52
::GUS in 4-day-old seedlings cultured under SDs; B:
Enlarged view indicated by blank rectangle in panel A; C:
Three-week-old plants cultured under short day photoperiods;
D: Inflorescence; E and F: Flowers at stage 12 and 15,
respectively; G: Silique; H: Enlarged view indicated by white
rectangle in panel G
1.3 Interaction between AGB1 and site-directed
mutants of AtPP2C52
Three site-directed mutants of AtPP2C52 (AtPP2C-
52
G99D
, AtPP2C52
G105D
and AtPP2C52
DGH102-104ERN
)
were generated (Figure 3A). These mutated sites were
highly conserved, and they are involved in the PP2C
active site (Das et al., 1996; Sheen, 1998). The
mutated sequences encoded unrelated amino acids.
None of these mutations affected the molecular weight
of these mutant proteins (Figure 3B). However, all of
these mutations abolished the interaction between
AGB1 and AtPP2C52 (Figure 3C).

