Basic HTML Version

International Journal of Marine Science 2014, Vol.4, No.50, 1-22
http://ijms.biopublisher.ca
9
was the only target gene that was affected by
temperature in both experiments, discounting its
utility as a housekeeping gene for this coral species.
However, the difference in expression between
samples of the two temperature regimes was only
~30% in the VTE.
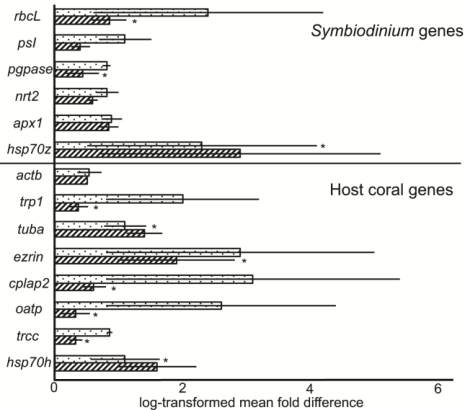
Figure 6 Global site of origin differences in gene expression. To
compare the average expression of each of the 14 target genes
across the two sites of origin, Houbihu (upwelling site [UWS];
speckled columns) and Houwan (non-upwelling site [NUWS];
hatched columns), data from the 12 UWS samples of the
Seriatopora hystrix
variable temperature experiment (VTE) and
all 24 samples of the elevated temperature experiment (ETE)
were compared against the expression data from the 12 NUWS
samples from the VTE. Please see the text or Tables A1-3 for
full gene names. Gene expression values were first converted to
fold changes relative to the lowest expression sample within the
48 assessed. Then, mean fold changes were log-transformed in
order to present all data on the same scale. Asterisks (“*”)
denote significant differences detected by Bonferroni-adjusted
Wilcoxon rank-sum tests, and error bars represent standard
error of the mean (
n
=36 and 12 for the UWS and NUWS,
respectively). The statistically significant difference in host
coral
oatp
expression between sites must be interpreted with
caution, as different primer concentrations were used between
experiments (Table 4). “
hsp70z
” and “
hsp70h
” refer to the
hsp70
homolog from the
Symbiodinium
and host coral
compartments, respectively.
This absence of a temperature-driven response in
12/14 target genes across both compartments of the
holobiont in samples exposed to 30
℃
may suggest
that exposure to a stable, elevated temperature regime
does not elicit a stress response in specimens of
S.
hystrix
from Southern Taiwan. Exposure to 30
℃
has
typically been shown to result in stress in
S. hystrix
elsewhere (Hoegh-Guldberg and Smith, 1989; Loya et
al., 2001), and this should manifest at the gene level
over the course of only several hours (Feder, 1996).
Although it is tempting to speculate that this lack of
an mRNA-level response to exposure to 30
℃
in none
of the six
Symbiodinium
target genes, and all but two
(
actb
and
tuba
) of the eight host coral gene targets,
stems from adaptations to life in a fluctuating
temperature environment (Mayfield et al., 2011), a
targeting of a greater number of gene and protein
candidates over a longer sampling time will be
necessary to demonstrate that these corals indeed
display no signs of a sub-cellular stress response upon
exposure to this temperature; indeed, <0.1% of the
transcriptome of the
S. hystrix
-
Symbiodinium
holobiont was queried herein, and current efforts
employing next generation sequencing-based
technology (Mayfield et al., in prep.) seek to assess
the whole-transcriptome response of these samples to
gain further insight into the molecular mechanisms by
which
S. hystrix
acclimates to altered temperature
regimes in the laboratory and acclimatizes to them in
the upwelling reef ecosystems in which they reside.
Additionally, in order to determine whether the
absence of a gene-level is related to thermal history, a
static ETE would also need to be conducted with
conspecifics from the NUWS; such an experiment
would help to strengthen the hypothesis put forth in
prior works (e.g., Oliver and Palumbi, 2011; Mayfield
et al., 2013b) that corals from variable temperature
environments perform better at elevated temperatures
than those from stable temperature habitats. If such a
differential response were indeed documented, it
would be attributable to acclimatization rather than
adaptation, as the UWS and NUWS populations were
found to be genetically identical (Table 2) and possess
similarly homogenous
Symbiodinium
assemblages.
That said, neutral markers, such as the microsatellites
employed herein, cannot detect genetic differences
driven by environmental changes (Foret et al., 2007)
and so future studies seeking to uncover genetic
differences underlying phenotypic plasticity should
not only utilize larger sample sizes but also analyze
genes that could be hypothesized to be under selection.

