Basic HTML Version


International Journal of Aquaculture 2012, Vol.2, No.2, 5
-
10
http://ija.sophiapublisher.com
8
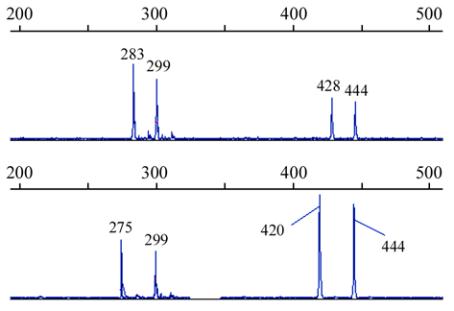
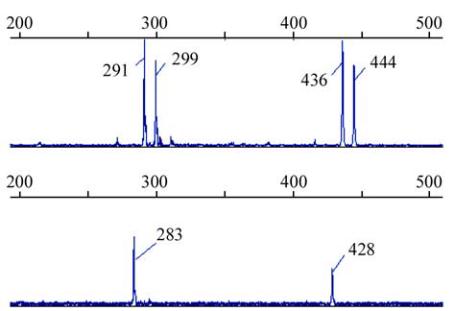
Figure 1 PCR amplifying fragments of duplicated loci
HUBA02
in 4
Babylonia areolata
individuals shown by GeneMapper 3.5
software
3 Materials and Methods
3.1 Samples and DNA extraction
Thirty-two
B. areolata
individuals were collected
from a wild population at the seashore near Qiaogang
town of Beihai city, Guanxi Province, China.
DNA
was extracted from the ethanol preserved
gastropod
muscles
using the Cell/Tissue Genomic DNA
Extraction Kit (TianGen, Beijing, China).
3.2 Microsatellite-enriched library construction and
sequencing
A microsatellite-enriched DNA library was constructed
using a selective hybridization and magnetic bead
enrichment protocol as described in Wang et al
(2010a). Briefly, about 38 µg genomic DNA of three
individuals was digested by the restriction enzyme
Mbo
I (TaKaRa, Dalian, China). Fragments of
300~1 000 bp collected with the Gel DNA Extraction
Kit (TianGen) were ligated to double stranded
Mbo
I
adapters by incubating with T4 DNA ligase (TaKaRa)
at 16
℃
overnight. Excess adapters were removed by
washing with 0.1×TE buffer (pH 8.0) on an Ultrafree
column (Pall, CA, USA). DNA fragments with
adapters were amplified with linker B for 5 PCR
cycles and purified using an Ultrafree column (Pall).
The amplified products were denatured and then
hybridized to biotin-labeled (CA)
12
, (GA)
12
, (ACA)
8
,
(AGA)
8
, (GACA)
6
and (GATA)
6
oligonucleotides
(mixed in advance at the ratio of 3:1:1:1:2:2, total 150
pmol) in 0.5×SSC at 68
℃
for 60 min. Washed four
times in 0.1×SSC at room temperature, DNA
fragments bound to these probes were then captured
with Streptavidin MagneSphere® Paramagnetic
Particles (Promega, USA) and eluted by DNase-free
water. The microsatellite-enriched elution was amplified
and purified as described above. The amplified
products were cloned using the T-Easy system
(Promega). Clones containing potential microsatellite
loci were selected and sequenced on ABI 3730xl DNA
analyzers at Sangon Biological Engineering Technology
& Services Co., Ltd. (Shanghai, China).
3.3 Primer design and genotyping
DNA sequences removed the linkers and T vector
were searched against each other and against 17
spotted babylon microsatellite sequences from
GenBank (accessed March 19, 2012) using Vector
NTI Advance 11.0.0 (http://www.invitrogen.com) to
check for duplicates. Microsatellite sequences containing
at least 6 di-, 5 tri-, 5 tetra-, 4 penta-, and 3 hexa-,
hepta- and octanucleotide repeats were selected using
MISA software (http://pgrc.ipk-gatersleben.de/misa/).
Good sequences with sufficient flanking regions were
used for primer design with Primer3 (http://biotools.
umassmed.edu/bioapps/primer3_www.cgi). An M13
(
-
21) universal leading sequence (5
-
TGTAAAACGA
CGGCCAGT
-
3) was added to the 5' end of each
forward primer (Schuelke, 2000), and primers were
synthesized by Sangon Biological Engineering
Technology & Services Co., Ltd (Shanghai, China).
All primer pairs were tested for PCR amplification in
the 32 wild individuals. PCR was conducted in a 10
mL solution containing about 30 ng template DNA,
and reagents (TianGen) as follows: 1× reaction buffer
containing 20 mM Tris-Hcl (pH 8.4), 20 mM KCl and
10 mM (NH
4
)
2
SO
4
, 1.2~2.0 mM MgCl
2
, 0.2 mM each
dNTP, 0.4 units Taq polymerase, 0.2 pmol M13 (
-
21)
tailed forward primer, 0.6 pmol reverse primer, 0.6
pmol M13 (
-
21) primer labeled with fluorescent dyes
(FAM, VIC, NED or PET, Applied Biosystems, Foster

