![]()
Genomics and Applied Biology 2015, Vol. 6, No. 2, 1-7
http://gab.biopublisher.ca
6
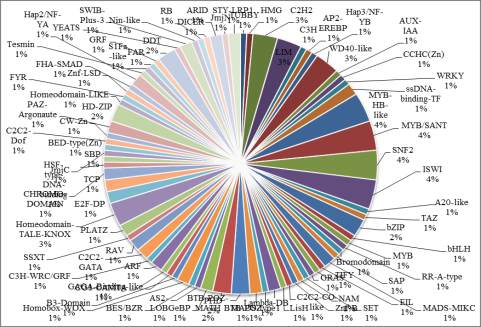
Figure 5 Plant Transcription Factor Result
a small fraction of tetra-, penta- and hexa-nucleotide
SSRs were identified in transcripts, the number is
quite significant.
5. Plant Transcription Factor
Further, transcription factor encoding transcripts were
identified by sequence comparison to known
transcription factor gene families. Result shows that
transcription factor genes distributed with at least 82
families were identified (Figure 5). The overall
distribution of transcription factor encoding transcripts
among the various known protein families is very
similar with that of other legumes as predicted earlier
(Libault et al., 2009).
Conclusions
This study is focus on
Vicia sativa
L. species
(SRR403901)
from NCBI database for de novo
Transcriptome
analysis
by
RNA-seq
using
next-generation Illumina sequencing. The transcriptome
Table 5. Statistics of SSRs identified in transcripts
SSR Mining:
Total number of sequences examined:
22748
Total size of examined sequences (bp): 11444673
Total number of identified SSRs: 1150
Number of SSR containing sequences: 1055
Number of sequences containing more than one
SSR:
92
Number of SSRs present in compound formation: 48
Distribution to different repeat type classes:
Mono-nucleotide
362
Di-nucleotide
243
Tri-nucleotide
529
Tetra-nucleotide
10
Penta-nucleotide
3
Hexa-nucleotide
3
sequencing enables various functional genomics